Selenoproteins
Iodothyronine deiodinase family (DIO)
The iodothyronine deodinase (DI) family of selenoproteins is constituted by three paralogous proteins in mammals (DI1, DI2 and DI3), which are involved in regulation of the thyroid hormone activity by reductive deodination8. The main thyroid hormone which is produced by thyroid is secreted in the inactive form, thyroxine (T4). These proteins’ family metabolize T4 to inactivate or deactivate it. Surprisingly, homologs of mammalian deodinases occur not only in other vertebrates, they are also found in simple eukaryotes and bacteria. The function of deodinase homologs in these organisms is unknown. These proteins have different subcellular localizations and tissue expression. All of them are transmembrane proteins which have a thioredoxin fold and form homodimers. The circulating levels of thyroid hormone are basically regulated by DI1 activity. However, DI2 and DI3 have been implicated in fine-tuning local intracellular T3 concentrations in a tissue-specific manner, without changing overall serum levels of T3. All of the iodothyronine deiodinases contain Sec. The active-site Sec residue is located in the N-terminal part of the protein. However, in DI2, an additional Sec, whose function is unknown, is present in the C-terminal region. Other authors state that it appears that the primary function of the second UGA is to serve as stop codon. This second Sec does not participate in the catalytic mechanism and is dispensable for the D2 functional activity. The 3 DIs have a low sequence identity (50%), but they have similar overall topology and structural organization12. The three Iodothyronine deiodinases (DI1, DI2 and DI3) are found in all the vertebrates.
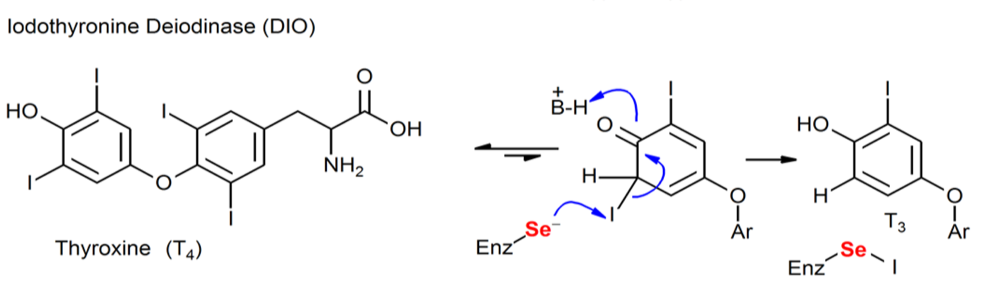
Figure. Chemical processes mediated by Sec-containing enzymes (DIO).
DI1 is a protein present in humans and DIO1 in lizard, but they are very similar so we analyse them together. As we can see, this protein is present in both species and they present very close results: all hits are genes situated in the SJPD01000026.1 scaffold. We obtain 1 hit in human, between positions 4652779-4652928, and 2 hits when comparing with the lizard between 4652788-465385. It is important to mention that in both cases, the alignment doesn’t start with a Methionine, there’s a huge gap in the beginning of the alignment, so we thought that maybe the exonerate programme has not correctly predicted the structure of the gene or maybe the first part has been lost during the evolution of the Selenoprotein.
Looking into the phylogenetic tree, we observe that this scaffolds we have mentioned before are related to the human and lizard’s queries used to compare with our specie. In this point, as the lizard contains the protein we are analysing, we are going to take into account the results obtained comparing with this specie, because is more relative to the Komodo in terms of evolution. We observe that it is a 2 exons gene. The T-coffee results were so good and the Seblastian’s ones too, where we could observe that the Selenocysteine is located in the second exon between 4653651 - 4653850, and because the gene is situated in the forward chain, the SECIS element (grade A) is located in the 3’UTR end between positions 4656858 - 4656927.
To sum up, we can conclude that the Selenocysteine is conserved and the Selenoprotein as well, and in addition to the SECIS elements we have found, we can say that the DI1/DIO1 protein is conserved in the Komodo.
DI2 is a protein present in humans and DIO2 in lizard, but they are very similar so we analyse them together. As we can see, they present similar results: when comparing with human, we only obtain 2 hits in the SJPD01000003.1 scaffold between positions 39328645-39341228, and in the lizard’s comparison, we obtain 2 hits in the same scaffold as the human’s case between positions 39328645-39341228 but another one in the SJPD01000026.1 scaffold, between positions 4652791-465292.
Looking into the phylogenetic tree, we observe that the human’s protein comparison correspond correctly with the Komodo’s scaffold. In the lizard’s case, we can discard the SJPD01000003.1 scaffold, because it is far situated from the lizard’s query in the phylogenetic tree, so we will take into account only the other scaffold, which is next to the query we are analysing.
In this point, as the lizard contains the protein we are analysing, we are going to take into account the results obtained comparing with this specie, because is more relative to the Komodo in evolution. We found a gene with one exon, situated between 39328618 - 39328782. This alignment doesn’t start with a Methionine. This could be because exonerate has not correctly predicted the structure of the gene and, therefore, these residues are lacking (including the Met at the beginning of every protein). But, even with this variation, we obtain very good T-coffee results and a SECIS element (grade A) on the Seblastian’s ones, situated on the same chain between positions 39332712 - 39332781, so it is situated in the 3’UTR.
So, in conclusion, we can conclude that the Selenocysteine is conserved and the Selenoprotein as well, and in addition to information obtained from the Seblastian, we can say that the DI2/DIO2 protein is conserved in the Komodo.
DI3 is a protein present in humans but not in lizard. Looking into the obtained results and hits when performing the blast and comparing it with the Komodo’s genome, we obtain an alignment of two genes situated in different scaffolds, but looking into the phylogenetic tree, we can discard one of them because of the distance they present in evolution. So finally we choose the hit from the SJPD01000026.1 scaffold, situated between positions 4652761 - 4652928.
So when we compare the DI3 human protein with our specie, we observe that it corresponds to a gene with 2 exons (exon 1: 4652761 - 4652929 i exon 2: 4653651 - 4653844) situated on the forward chain. This alignment doesn’t start with a Methionine. This could be because exonerate has not correctly predicted the structure of the gene and, therefore, these residues are lacking. Eventhough, we obtain pretty good results on the T-coffee analysis, and looking into the Seblastian results, we observe that there is a SECIS element (grade A) situated on the 3’UTR end of the same chain as the gene, between 4656858 - 4656927. We do not know in which of the 2 exons is situated the Selenocysteine, but both of them are pretty near to the SECIS element.
Now, in theory we could be able to conclude that the Komodo has the DI3 protein. But, taking into account the species evolution, we cannot conclude this because the lizard, which is a ancestor more near to the Komodo than the human, doesn’t have the DI3 protein, and it may have lost it due to evolutionary divergence. To solve it, we have consulted the SelenoDB programme to see if another specie nearest to the Komodo than the Human conserve this Selenoprotein, for example the chicken (Gallus gallus). In this case, the chicken has the DI3 protein, so we decided to check if there is a good alignment with this protein.
In this case, we obtained similar results as before: a 2 exon gene on the forward chain and a Selenocysteine aligned in the T-coffee situated in the second exon, between positions 52459661 - 52460551 and a SECIS element (grade A) in the 3’UTR end situated between positions 52461174 - 52461253. Now, we are able to perform conclusions and say that he Selenocysteine is conserved and the Selenoprotein as well, so we can say that the DI3/DIO3 protein is conserved in the Komodo.
Glutathione peroxidases (GPx)
Glutathione peroxidases are the largest selenoprotein family in vertebrates, and in addition, they are found in single-cell eukaryotes and prokaryotes. GPxs play physiological functions in organisms involved in hydrogen peroxide (H2O2) signaling, detoxification of hydroperoxides and maintaining cellular redox homeostasis. Selenoproteins of the glutathione peroxidase (GPx) family are widespread in all three domains of life. In mammals, there are eight GPx paralogs, from which five (GPx1, GPx2, GPx3, GPx4, and GPx6) contain a Sec residue in their active site. In the other three GPx homologs (GPx5, GPx7, and GPx8), the active-site Sec is replaced by Cys. Moreover, GPx6 homologs in some mammals are not selenoproteins and have a Cys in the active site8. They all are highly conserved, and this is even more evident in Sec-containing GPx (in the case of mammalian GPx, they share approximately 80% of identity)12. GPx1, GPx2, GPx3 and possibly GPx6 work as tetramers, whereas GPx4 is a monomer. This protein family includes the two types of selenoproteins mentioned in the introduction of this website. GPx1 is a stress-related selenoprotein which is highly regulated by Selenium availability (its expression decreases drastically when there is lack of Selenium), whereas GPx4 is a housekeeping selenoprotein (it is less affected by dietary Selenium status and often serve functions critical to cell survival)8.
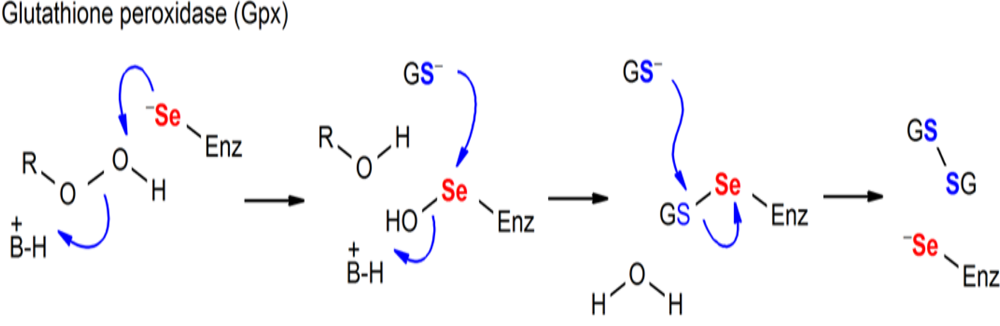
Figure. Chemical processes mediated by Sec-containing enzymes (GPx).
GPx1 is a protein present in humans and in lizard. They present similar results and hits when performing the blast and comparing it with the Komodo’s genome. We found huge amount of scaffolds in both species, this is not rare because, as we know, there are lots of proteins in this family and they are very similar to each other.
To choose the most interesting hits, we look into the phylogenetic tree. In the human’s comparison, the most relative hit to the original human protein was the ones located in the SJPD01000040.1 scaffold, between positions 929848-930189, and in the lizard’s case, it was the ones situated on the same scaffold, between positions 92663 - 93018.
So, as the lizard has got the protein we are analysing, we are going to take into account the results obtained comparing with this specie, because is more relative to the Komodo into phylogenetic terms. We observe that it corresponds to a gene with 2 exons situated on the forward chain. This alignment doesn’t start with a Methionine. This could be because exonerate has not correctly predicted the structure of the gene and, therefore, these residues are lacking (including the Met at the beginning of every protein). Eventhough, we obtain pretty good results on the T-coffee analysis, and looking into the Seblastian results, we observe that there is a SECIS element (grade A) situated on the 3’UTR end of the same chain as the gene, between 93235 - 93299, and the predicted Selenocysteine is located between positions 89436 - 89657, on the first exon of the gene.
To sum up, we can conclude that the Selenocysteine is conserved in the lizard and the Selenoprotein as well, and in addition to the SECIS elements we have found, we can say that the GPx1 protein is conserved in the Komodo.
GPx2 is a protein present in humans and in lizard and they present similar results. GPx2 is very similar to the GPx1’s case, so their results and analysis will be very similar as well. To choose the most interesting hits, we look into the phylogenetic tree. In the human’s comparison, the most relative hit to the original human protein was the ones located in the SJPD01000040.1 scaffold, between positions 926621 - 930195, and in the lizard’s case, it was the ones situated on the same scaffold, between positions 926621 - 93019.
So, as the lizard contains the protein we are analysing, we are going to take into account the results obtained comparing with this specie, because is more relative to the Komodo in evolution. We observe that it corresponds to a gene with 2 exons situated on the forward chain. This alignment starts with a Methionine, meaning that probably we have aligned the entire real protein. So, we obtain very good results on the T-coffee analysis, and looking into the Seblastian results, we observe that there is a SECIS element (grade B) situated on the 3’UTR end of the same chain as the gene, between 930420 - 930484, and the predicted Selenocysteine is located between 926621 - 926842, on the first exon.
In conclusion, we can say that the Selenocysteine is conserved in the lizard and the Selenoprotein as well, and in addition to the SECIS elements we have found, we can say that the GPx2 protein is conserved in the Komodo.
GPx3 is a protein present in humans and in lizard and they present similar results. It is very similar to the all the GPx’s cases we have seen before. To choose the most interesting hits, we look into the phylogenetic tree. In the human’s comparison, the most relative hit to the original human protein was the ones located in the SJPD01000009.1 scaffold, between positions 1771450 - 17721381, and in the lizard’s case, it was the ones situated on the same scaffold, between positions 17714506 - 17714506.
So, as the lizard contains the protein we are analysing, we are going to take into account the results obtained comparing with this specie, because is more relative to the Komodo in evolution. We observe that it corresponds to a gene with 4 exons situated on the forward chain. This alignment doesn’t start with a Methionine. This could be because exonerate has not correctly predicted the structure of the gene and, therefore, these residues are lacking (including the Met at the beginning of every protein). Eventhough, we obtain pretty good results on the T-coffee analysis, and looking into the Seblastian results, we observe that there are no SECIS elements in this case. It is important to mention that, when doing the comparison with the human query, we obtain the same negative results. In this point, we tried to analyse the other hit we obtained in the lizard’s case, because maybe this results are due to a truncation, but the other possible scaffold obtained with the t-blast was situated in the other chain, the reverse chain, meaning that this fact is not caused by a division of our protein.
In conclusion, we cannot say that there is a Selenocysteine conserved neither in the human and the lizard, because even though we found a positive Selenocysteine alignment in the T-coffee results, we were not able to found relative SECIS elements in this position, so to summarise, we cannot confirm that the GPx3 protein is conserved in the Komodo. More research should be done in order to confirm this hypothesis, because it could be an error of the Seblastian and the SECISSearch programs.
GPx4 is a protein present in humans and in lizard and they present similar results. In this case, we only obtained 1 hit per specie, so it wasn’t necessary to use the phylogenetic tree to choose the correct scaffolds. When comparing with the human protein, the gene was located in the SJPD01000092.1 scaffold, between positions 2048178 - 2048609, and when comparing with the lizard protein, the gene was located on the same scaffold, between positions 2048178 - 2048339.
First, we started analysing the lizard’s results, because is more relative to the Komodo in evolution. We observed that the gene was situated on the reverse chain and it had 2 exons, but, when looking into the T-coffee results, there was no Selenocysteine alignment. This could mean that the Selenocysteine residue changed to a Cysteine during evolution, or maybe that the lizard’s query was not well annotated on the SelenoDB database. To check that, we observed that the original lizard’s query did not contain any Selenocysteine or Cysteine neither. In this point, we changed of point of view and started analysing the results obtained from the human’s comparison. Here we saw that the gene was situated on the reverse chain and it had 4 exons and a good T-coffee alignment. Then, we performed a Seblastian test, and we found that there iwa a SECIS element (grade A) in the 3’UTR end of the same chain as our gene was found, between positions 2046121 - 2046047, and a possible Selenocysteine between positions 2048463 - 2048607.
In order to clarify our results, we tried to analyse the same protein but from the chicken, but it is not annotated on the SelenoDB database. So finally, we discarted the lizard’s query because we think that the region where the Selenocysteine must be located is not found in the sequence taken from our database, so the lizard protein sequence used for the prediction was incomplete from the beginning. But, taking into account the results obtained when comparing with the human protein, we can conclude that the Selenocysteine is conserved in the lizard and the Selenoprotein as well, and in addition to the SECIS elements we have found, we can say that the GPx4 protein is conserved in the Komodo.
GPx5 is a protein present in humans but not in lizard. So, our hypothesis is that it is not very probable that Komodo has it since, in terms of evolution, it is more near to the lizard than to the human. To solve this problem, we tried to find out if the chicken had this protein, but we could not find the GPx5 protein on the SelenoDB. In addition, comment that the hits found were in scaffolds also present in other GPx proteins, with better identity, so we think that the results obtained were just because this protein is very similar to others of the same family, instead of because the snake has this protein. And, as the chicken doesn’t have this protein as well, it can be concluded that the common ancestor of the lizard, the chicken and the Komodo lost this duplication or it did not win it whereas the ancestor of the human did.
GPx6 is a protein present in humans but not in lizard, like in the case before. So, our hypothesis is the same as with the GPx5, is that it is not very probable that Komodo has it since, in terms of evolution, it is more near to the lizard than to the human. As we have said before, we tried to find out if the chicken had this protein, but we could not find it on the SelenoDB. So, as the chicken doesn’t have this protein as well, it can be concluded that the common ancestor of the lizard, the chicken and the Komodo lost this duplication or it did not win it whereas the ancestor of the human did.
GPx7 is a protein present in humans but not in lizard, like in the case before. So, our hypothesis is the same as with the GPx5 and GPx6, is that it is not very probable that Komodo has it since, in terms of evolution, it is more near to the lizard than to the human. As we have said before, we tried to find out if the chicken had this protein, and in this case we found it.
When performing the comparison with both species, we obtain hits on the same scaffolds for all of them, but no Selenocysteine alignment is found in any of the hits analysed. In this point we could think that the Selenocysteine evolved to a Cys-containing homolog. To confirm this hypothesis, we performed a Seblastian/SECISSearch analysis to find out if there are SECIS elements near our gene is located. Unfortunately, we could not find any relevant SECIS in human and neither in the chicken.
So, in conclusion and taking into account the previous results, we can say that the GPx7 protein is not included in the Komodo’s genome.
GPx8 is a protein present in humans and in lizard and they present similar results and hits when performing the blast and comparing it with the Komodo’s genome. To choose the most interesting hits, we look into the phylogenetic tree. In the human’s case, the most relative hit to the original human protein was the ones located in the SJPD01000005.1 scaffold, between 4298996-4300345, and in the lizard’s case, it was the ones situated on the same scaffold, between 4300082-4300345.
So, as the lizard contains the protein we are analysing, we are going to take into account the results obtained comparing with this specie, because is more relative to the Komodo in evolution. We observe that it corresponds to a gene with 2 exons situated on the forward chain. This alignment doesn’t start with a Methionine. This could be because exonerate has not correctly predicted the structure of the gene and, therefore, these residues are lacking (including the Met at the beginning of every protein). In addition, we did not found a positive Selenocysteine alignment in the T-coffee results. In this point we could think that the Selenocysteine could have evolved to a Cysteine (Cys-containing homolog). To confirm this hypothesis, we performed a Seblastian/SECISSearch analysis to find out if there are SECIS elements near our gene is located. Unfortunately, we could not find any relevant SECIS in lizard, because they were situated too far away from our gene. In this point we thought that it could be interesting to use the GPX8 chicken protein to perform the analysis, but as the other cases, any relevant SECIS were found.
So, in conclusion and taking into account the previous results, we cannot confirm that the GPx8 protein is included in the Komodo.
We present here an unambiguous phylogeny of the GPx tree wherein three evolutionary groups were observed: GPx1/GPx2, GPx3/GPx5/GPx6, and GPx4/GPx7/GPx8. It appeared that Cys-containing GPx7 and GPx8 evolved from a GPx4-like selenoprotein ancestor, but this happened prior to separation of mammals and fishes. GPx5 and GPx6 are the most recently evolved GPxs, which appeared to be the result of a tandem duplication of GPx3 at the root of placental mammals. Interestingly, no Sec-containing GPx5 form could be identified. As phylogeny indicates that this protein evolved from a duplication of selenoprotein GPx3, the Sec to Cys displacement must have happened very early in the evolution of GPx5.
Methionine sulfoxide reductase (MSRA)
MsrA is a sulfoxide reductase, present in humans and some eukaryotic species. As the Methionine amino acid is highly susceptible to oxidation, it ends up being a mixt of Methionine-S-sulfoxide and Methionine-R-sulfoxide. Its function, complementary to MsrB1, is to reduce the Methionine-S-sulfoxide levels8. However, MsrA and MsrB have different structures and belong to different Selenoprotein families.
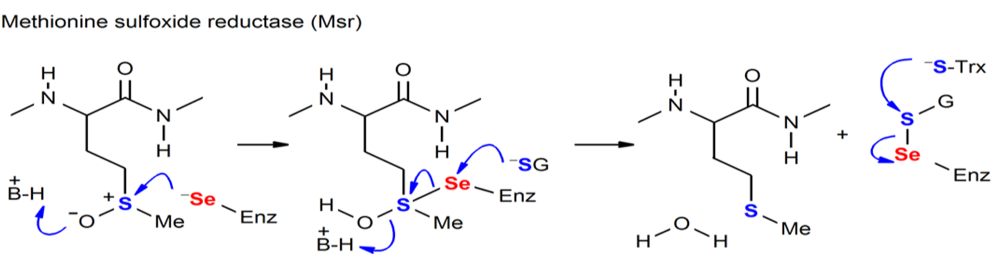
Figure. Chemical processes mediated by Sec-containing enzymes (MSR).
MsrA is a protein present in humans and in lizard. They present similar results and hits when performing the blast and comparing it with the Komodo’s genome. When performing the Tblast, we obtained hits in 2 different scaffolds (the same ones for both species). To choose the most interesting hits, we look into the phylogenetic tree. In the human’s comparison, the most relative hit to the original human protein was the ones located in the SJPD01000076.1 scaffold, between positions 149906 - 162828, and in the lizard’s case, it was the ones situated on the same scaffold, between positions 149907 - 164533.
So, as the lizard has got the protein we are analysing, we are going to take into account the results obtained comparing with this specie, because is more relative to the Komodo into phylogenetic terms. We observe that it corresponds to a gene with 5 exons situated on the reverse chain. This alignment doesn’t start with a Methionine. This could be because exonerate has not correctly predicted the structure of the gene and, therefore, these residues are lacking (including the Met at the beginning of every protein). Moreover, there is not a Selenocysteine aligned in the T-coffee results, so our next step was to do a Seblastian/SECISearch analysis, to detect if there are SECIS elements related to that gene, in order to predict if it could be a Cys-containing homolog, but we did not obtained valid positive SECIS elements related to our scaffolds.
So, in conclusion and taking into account the previous results, we cannot confirm that the MsrA protein is included in the Komodo, because of the T-coffee and the Seblastian results.
Selenoprotein 15 (Sel15)
This protein has a thioredoxin-like domain and contains an NTD signal peptide, consistent with its ER localization. Its function is the reduction or rearrangement of disulfide bonds in the ER-localized or secretory proteins8.
This protein is found in the lizard and human. When running the blast, hits are found in the scaffold SJPD01000031.1 between positions 5149003-5148833 and 5149003-5115475 in human and lizard respectively. In the case of the human, the protein predicted has 2 exons in the reverse strand, whereas in the case of the lizard the protein predicted has 4 exons also in the reverse strand. Both T-coffees present very high scores, but in the case of the human, there is a gap at the beginning and at the end, meaning that part of the protein is missing. However, in the case of the lizard, there are just a few amino acid changes and no gaps. In addition, the selenocysteine found in the predicted protein matches with the Selenocysteine in both queries. Regarding the results of the Seblastian, significant SECIS elements are found in 3’ UTR region (between positions 5108382-5108309 in human and 5115024-5114951 in lizard).
Therefore, we can conclude that Sel15 is a selenoprotein in Varanus komodoensis.
Selenoprotein I (SelI)
Selenoprotein I catalyzes phosphatidylethanolamine biosynthesis and plays an important role in the formation and maintenance of the membranes of the vesicles7.
In its analysis we found the same significant scaffold in human and lizard, SJPD010000126. Human’s positions were 951142 – 964258, in the reverse strand and containing 9 exons. Lizard’s ones were 960906-962118, also in the reverse strand and containing, this time, 10 exons. Both t coffee scores were high, 999 and 997, but only in the human predicted protein we had aligned X. In the Lizard results, the first part of the predicted protein was missing, probably because it was in another scaffold.
For this last reason, and thanks to the fact that selenoprotein had multiple isoforms in lizard, we performed another analysis. We chose, from this other isoform, the scaffold SJPD01000126, and the positions from 951124-951318. The resultant region had 3 exons, in the reverse strand, and a t coffee score of 995. X were aligned in the results this time so we could validate this selenoprotein’s presence by both species analysis.
After running the Seblastian, Selenoprotein I was predicted both in the human comparation and in the chosen isoform in the Lizard. With all of that, we could conclude that Varanus komodoensis has Selenoprotein I.
Selenoprotein K (SelK)
Selenoprotein K is involved in calcium flux in immune cells, T cell proliferation and neutrophil migration. It is also related with the endoplasmatic reticulum-associated degradation of glycosylated proteins and plays a key role in the protection of cells from the apoptosis induced by the ER stress11.
This protein is also found in both species and although many hits were found in many scaffolds when performing the blast, only one hit of each specie was left after the selection criteria. The scaffold which contained the hits was the same for the two of them, the SJPD01000007. Human’s hit was in positions 14408200 - 14408295 and lizard’s one in 14408200 – 14408295, both in the forward strand, containing 4 exons and starting with methionine. T coffee score was not especially high in human, being a 989, but it was pretty high in Lizard, with a 994. Analysing the results of the T coffee we see that in human there’s an X but it is not aligned in the predicted protein: three residues are lost in the same positions where in human we have a Sec. But then, when analysing the Lizard’s T coffee, we see that there’s an X aligned with another X. This makes us think that the Selenoprotein K exists in the Varanus komodoensis.
As we only found a relevant X alignment with the Lizard, we performed its Seblastian analysis and found one SECIS. We checked the position and saw it corresponded with what we predicted. Again, with that, we confirm the selenoprotein nature of SelK in Varanus komodoensis, although in the human comparison we didn’t find the Sec for the predicted protein probably due to its further phylogenetic distance with our specie.
Selenoprotein S (SelS)
As SelK, the protein Sel S is involved in the transfer of misfolded proteins from endoplasmatic reticulum to the cytosol. That means it is involved in recognition, ubiquitination, and retrotranslocation of protein substrates from the ER to the cytosol, and their subsequent degradation by the ubiquitin/proteasome.
This protein is found in both species. However, when we run the blast for human, no output is shown, meaning that any hit presents the conditions established (identity >50% and e-value <0,00001). However, when running the blast for the lizard genome, two significant hits were found between the positions 5580804-5582539 in the SJD01000057.1 scaffold. The T-coffee shows a perfect alignment (score = 1000), covering almost the entire protein sequence. In the predicted protein, 5 exons were found in the reverse strand. Thus, we can confirm that Selenoprotein S exists in Varanus Komodoensis genome.
In the T-coffee results, it can also be seen that Selenocysteine is conserved and the Seblastian results indicate the presence of a SECIS element in the reverse strand in 3’UTR (positions 5575992-5575917). Therefore, we can conclude that SelS is a selenoprotein in Varanus Komodoensis.
Selenoprotein M (SelM)
Selenoprotein M is a thiol-disulfide oxidoreductase that resides in the ER. It is highly expressed in the brain and has important neuroprotective properties. It has active site that consist of a selenocysteine-containing thioredoxin-like domain, that mediates thiol-disulfide exchange8.
As it has happened many times, we found many hits in different scaffolds in both species, but again, only one scaffold had hits that overcame the filtering criteria. The scaffold’s name is SJPD01000053 and it contained one hit in human, from 3410972 – 3411133 and two hits in Lizard, which extended from 3410086 – 3411103, both in the forward strands. Predicted proteins had 3 exons and their T coffee scores were very high, 998 and 999, respectively. Analysing these results, we saw that, for both comparisons, there were X in our queries, in the first positions, but they were not aligned because the first and the last amino acids of our predicted proteins were missing. This may be due to a bad prediction of the exonerate program.
However, as we had Sec in our analysis queries, we performed the Seblastian. There we could predict two SECIS (one in each strand) in both species. We analyzed the ones in the forward chains. After checking the positions and confirming they were coherent with our exons, we can confirm the presence of SelM in Varanus komodoensis.
Selenoprotein N (SelN)
Selenoprotein N is a protein that plays a key role in cell protection against oxidative stress and in the regulation of the calcium homeostasis related to redox mechanisms. It regulates the calcium levels in the endoplasmic reticulum taking care of the good functionality of the calcium pump12.
A lot of hits were found when performing the blast, but almost all of them (in fact, all of them except one from Human) were in the same scaffold: SJPD01000036. Human’s chosen positions extended from 7844146 – 782210, including 10 exons. Lizard’s ones were from 7844146 – 7862222, containing 11 exons. Both were in the reverse strands. Performing the T coffee, we obtained very high scores: 992 and 1000, but in the human analysis a lot of residues were missing in the beginning of our predicted protein, probably due to exonerate. We found X aligned analyzing the results of both t coffee.
We corroborated the results by using Seblastian and they showed us three SECIS elements, for both species. We discarded the ones in the positive strands, and we chose the ones which positions were close to our predicted protein’s exons.
For all of that, we can accept our prediction and confirm Selenoprotein N is in Varanus komodoensis.
Selenoprotein O (SelO)
Selenoprotein O is a protein that’s in charge of catalyzing the transfer of AMP to Ser, Thr and Tyr residues of target proteins (AMPylation). It is not well characterized but is thought to be a redox-active mitochondrial selenoprotein which interacts with redox target protein12.
Many hits were found in both comparations, but after filtering, we had only one scaffold with many hits in human and two significant scaffolds, with different hits, in lizard. The region predicted in human was in the reverse strand, scaffold SJPD01000016, extending from the positions 1978997 – 1986356 and had 10 exons. Its T coffee score was of 992, and it had 1 aligned X. In the comparation with the Lizard, we analyzed both filtered scaffolds, SJDP01000016 from the 1984204 to 1986122 and SJDP01000020, from 22000032 to 22009673. First region had 2 exons and the second one had 8, and they both had high T coffee scores. Unfortunately, no X were found in the alignments and no cysteines were aligned either and some residues of the predicted proteins were missing. However, this Selenoprotein O has an isoform in Lizard, so we analyzed this other query. In this one we also found many hits but only one scaffold, with two hits, overcame the filtering. It was SJDP01000020 and the region of interest 22025489 – 22025904. It had 2 exons and a 1000 score in T coffee. Again, no X were in the alignment but this time we found aligned cysteines. Even with that, we couldn’t conclude it was a cysteine homologous because in the initial query there were no X so maybe the aligned cysteines are just normal cysteines.
Performing the Seblastian for the human comparation we saw it predicted selenoprotein O. It is not how we expected it to be as lizard should be more similar to our specie, but maybe we found the alignment only between our query and the human genome because it’s better annotated than the lizard one. Again, with this, we can conclude that Selenoprotein O is found in Varanus komodoensis.
Selenoprotein P (SelP)
Selenoprotein P is a protein that contains more than one selenocysteine residue. It is a secreted protein, formed by two domains, that contains the majority of the selenium in plasma. Its function is not well known but it might have an extracellular oxidant defense role because its presence is correlated with selenium protection8.
Many hits were found in different scaffolds in both comparations. In human, after filtering, there was only a scaffold left with three different hits. The scaffold was SJDP01000005 and the studied sequence 747511 – 749227. It had three exons, located in the reverse strand. T coffee score was really high, 996, the protein predicted started with a methionine and there was an aligned X. With the Seblastian, we found SECIS and a selenoprotein.
Lizard had two isoforms for Selenoprotein P, and many scaffolds that overcame the filtering criteria. We chose one scaffold from each isoform based on its high t coffee scores and the presence of aligned X. In both cases, the scaffold chosen is the SJPD100000005. First isoform’s region has 3 exons, from 747511-747633. The second one has also 3 exons, and it’s from 747508-749227. Both of them had, as it has just been said, aligned X, and when performing the Seblastian, it found the selenoprotein. With that, we can confirm by using the data conferred by both species comparations, that Varanus komodoensis has selenoprotein P.
Selenoprotein R or Methionine-sulfoxide reductase (SelR or MSRB)
Selenoprotein R is known as Methionine-sulfoxide reductase and its function is to reduce methionine-sulfoxide back to methionine. It has three important isoforms, that we analyzed separately.

Figure. Chemical processes mediated by Sec-containing enzymes (MSR).
After filtering the blast results of MSRB1 we found significant hits in one scaffold, SJPD000085, in both species. Positions were from 1870578-1872496 for human, and 1870593-1872496 in lizard, both in the forward strands and having 2 exons each. T coffee scores were high, 992 and 1000, and both results showed aligned X. With the Seblastian we found 2 SECIS in each species, but no Selenoproteins. Having a look again at the results, we saw, in both cases, aligned cysteines. SECIS were found in the positive strand by using Seblastian and they could be selected by proximity to our predicted region. With all this we can confirm the presence of Methionine-R-sulfoxide reductase 1.
MSRB2 has two scaffolds of interest that have overcome the filtering criteria in human, and one that has done it in lizard. We have chosen to analyze the scaffold SJPD000020 in both cases. Human’s positions go from 23300656 – 23299803 and lizard’s one from 23300659 – 23299803, all of them in the reverse strands and having 4 exons each. T coffee scores were high, 999 and 1000, although in human the first part of the alignment was missing. No X were present in any of the cases, but there were many aligned cysteines.
After performing the Seblastian, we confirm no SECIS can be predicted, neither with the SECIS3 program. As there were no X in the initial query, we can’t confirm whether these cysteines come from a selenoprotein so we can’t say it is a cys homologous. For that reason, MSRB2 is not present in Varanus komodoensis.
MSRB3 had two significant scaffolds in the human analysis and one in the lizard analysis. We chose the same scaffold for both species’ analysis because, even one of the lizard’s scaffolds gave a higher t coffee score (995), the alignment lacked a lot of the first aminoacids. So, the scaffold studied was SJPD1000017, from positions 758769 – 805563. It had 4 exons, reverse strand, and a low t coffee score, but the alignment was good. It didn’t contain any X but it did contain a lot of aligned cysteines. Seblastian didn’t predict any SECIS.
Lizard’s chosen scaffold was the same, SJPD01000017, with its positions of interest going from 764575 – 819877. It had 6 exons, located in the reverse strand. T coffee score was of 1000, but no X were aligned. Again, there were many cysteines aligned and after performing the Seblastian and seeing that no SECIS were predicted. As there were no X in the initial query, we can’t confirm whether these cysteines come from a selenoprotein so we can’t say it is a cys homologous. For that reason, MSRB3 is not present in Varanus komodoensis.
Selenoproteins W, T, H and V
Selenoproteins W (SelW), T (SelT), H (SelH) and V (SelV) belong to the Rdx family of selenoproteins. The members of this protein family possess a thioredoxin-like fold and are characterized by the presence of a conserved Cys-x-x-Sec motif. The Rdx family proteins are thiol-based oxidoreductases, but the exact function of any of these proteins remains unknown8.
Selenoprotein W (SelW)
SelW is one of the first identified Sec-containing proteins and is one of the most abundant selenoproteins in mammals. SelW belongs to the stress-related group of selenoproteins as its expression is highly regulated by the availability of Se in the diet. While SelW1 can be found in human with the conserved selenocysteine residue, SelW2 was lost in all tetrapods (but frog), and its homolog can be found under the name Rdx12 with a cysteine residue instead. The hypothesis is that before the split of amphibians SelW2 duplicated and was immediately converted to a Cys form generating Rdx12, and then SelW2 was lost prior to the split of reptiles12.
This protein is found in both species, human and lizard. When the blast is run, one hit is obtained in the same scaffold in both species, SJPD01000036.1, between positions 11391124-11391029 and 11391103-11391029 in human and lizard respectively. Both protein predictions have 3 exons in the reverse strand. In addition, the results obtained through T-coffee present a very high score, but in the case of the human, there is a gap at the beginning meaning that a part of the protein has not been found and may be in another part of the genome. However, no other significant hits are found when running the blast, so we can not determine where is the lacking part of this protein.
Regarding the selenoproteins characteristics, the selenocysteine found in the human query is not aligned with any part of the predicted protein (because it is the part that is missing). Therefore, we can not determine if it is a selenoprotein or not. On the other hand, in the lizard predicted protein, the selenocysteine is not aligned with another selenocysteine. In the sequence studied, two SECIS were predicted. One grade A SECIS was found after the gene, in the 3’ UTR region, in the same strand. Although the komodo dragon protein might have a selenoprotein or not, the presence of the SECIS element can be explained by the fact that SelW1 is a selenoprotein in humans.
Above all, we can conclude that SelW1 is present in Varanus komodoensis. but we can not confirm if SelW1 is a selenoprotein or not.
This protein is only found in the human genome. The results of the blast show 3 significant hits with a high percentage of identity in the scaffold SJPD01000061.1 between positions 3248079-3250889. In the predicted protein, 3 exons are found in the reverse strand. The T-coffee score is 1000 but there is a 19 amino acid gap at the beginning. This can be because exonerate has not predicted the structure of the gene correctly, and these amino acids are lacking (including the methionine).
On the other hand, instead of selenocysteine, we can see that there are 3 cysteines found and all of them match with another cysteine in the Varanus komodoensis genome. It make sense, as it is known that SelW2 is not a selenoprotein in humans, but a Cys-containing homolog. Therefore, although we cannot say which of the cysteines was a selenocysteine in the past, we can confirm that SelW2 is not a selenoprotein in Varanus komodoensis. Also, this is the reason why Seblastian predicts one SECIS element in the 3’ UTR region (positions 3300230-3300161).
Therefore, we conclude that SelW2 is a Cys-containing homolog in Varanus komodoensis.
Selenoprotein T (SelT)
SelT belongs to the Rdx family of selenoproteins. The members of this protein family are characterized by the presence of a conserved Cysx-x-Sec motif12. This protein is predominantly localized to the ER and Golgi and is ubiquitously expressed both during embryonic development and in adult tissues8.It was proposed that the Rdx family proteins are thiol-based oxidoreductases, but the exact function of any of these proteins remains unknown12.
This protein is found in both species, human and lizard. The hits were all found in the same scaffold (SJPD01000032.1) between positions 13468766-13471916 in both cases. The predicted protein from the human presents 5 exons in the forward strand, whereas the protein predicted from the lizard presents 4 exons. Regarding the T-coffee results, we can see that both scores are very good. However, the one from the lizard alignment presents a perfect match (score = 1000). This is probably due to the fact that Varanus Komodoensis is more similar to the lizard than to the human.
Anyhow, the Selenocysteine is conserved in both cases and when running Seblastian, two SECIS elements are predicted also in both species. One of the SECIS is located in the reverse strand, so it is not valid. Therefore, we take into account only the SECIS element found in 3’ UTR positions (between positions 13472734-13472817 and 13472734-13472817 for human and lizard respectively).
Considering all this evidence, we can conclude that SelT is a selenoprotein found in Varanus Komodoensis.
Selenoprotein H (SelH)
Selenoprotein H is an oxidoreductase that plays an important role in neuron protection against UVB-induced damage, by inhibiting the pathways that lead to cell apoptosis, promoting the mitochondrial synthesis and function and suppressing cellular senescence through genome maintenance and regulation of redox reactions5.
This protein is found in both species, human and lizard, where many hits were found in many different scaffolds when performing the blasts. Only one hit in human and two hits in lizard overcame the cut-off criteria, all of them found in the same scaffold, SJPD01000122. The one from human was located in the positions 569175 – 569258 and the sequence studied from lizard went through the positions 569172 – 572138. T coffee scores were 980 in human and a perfect 1000 in Lizard, and both predicted proteins were found in the reverse strands. The one from human had three exons and started with a methionine, and the one from Lizard only had 2 exons. When analysing both T coffee results, aligned X were seen in both species, making us confirm the existence of Selenoprotein H in Varanus komodoensis.
Checking the Seblastian results, we found two SECIS in both species. We discarded the ones that were in the positive strand, and we saw that we had SECIS elements in the reverse strand in both species. We checked the position of these SECIS, and we corroborated that they were in an acceptable nucleotide distance. With all, we can definitely confirm the selenoprotein nature of SelH.
Selenoprotein V (SelV)
SelV recently evolved, most likely by duplication from SelW [Mariotti et al., 2012] and then it was modified by addition of N-terminal sequences, whose function is unclear. This protein is found only in placental mammals. However, it was specifically lost in some organisms including gorillas12.
This protein is only found in humans. However, when we run the blast, any prediction of a scaffold that could contain SelV in Varanus komodoensis is found. The reason is the same as in the previous case, probably the hits are not significant and the identities are lower than 50%. However, the results obtained make sense if we take into account the literature, because SelV is not supposed to be present in our organism.
Selenoprotein U (SelU)
In high mammalian species, such as humans and mice, all SelU proteins exist in Cys form, due to the Sec to Cys event that occurred in the early period of mammalian history for the SelU lineage. Three subfamilies of SelU family, SelU1, 2 and 3 are found in humans.The Prx-like2 structure domain presented in these proteins implies that they belong to the thioredoxin-like superfamily8.
This protein is present in both human and lizard. All the hits are found in the scaffold SJPD01000014.1 with high percentage of identity between positions 6965627-6970639 in human and 6965164-6972460 in lizard. Regarding the global alignment obtained through T-coffee, a perfect match can be seen in the case of the lizard (score = 1000), whereas in the case of the human the score is a bit lower (score = 992). In both species predicted proteins, 5 exons were found in the forward strand.
However, no Selenocysteine is found in the human genome, but a Cysteine instead. Still, in the lizard predicted protein, a Selenocysteine is aligned with another Selenocysteine. Moreover, when we do the Seblastian for the protein predicted from the lizard, one SECIS element is found in the 3’ UTR region (positions 6972642-6972712). Therefore, we can conclude that SelU1 is a selenoprotein in Varanus Komodoensis.
This protein is found in human and lizard. When running the blast, significant hits are obtained in the scaffold SJPD01000005.1. From the global alignment obtained through T-coffee a good alignment can be seen in both species. However, the protein predicted in both cases lacks the first 30 amino acids approximately. This fact can be due to a not correct predicted structure of the gene from exonerate. In both species predicted proteins, 6 exons were found in the forward strand.
Regarding the Selenoproteins characteristics, no Selenocysteine was aligned with another Selenocysteine either in human and lizard. Instead, the predicted protein contains 3 Cys in the same positions as human and lizard SelU2 proteins do. In addition, just one SECIS is predicted but in the reverse strand, so it is no valid.
Taking everything into account, we can confirm that SelU2 is a Cys-containing homolog in Varanus Komodoensis.
This protein only exists in the human proteome. However, when we run the blast, no output is obtained. This probably means that the hits are not significant and the identities are lower than 50%. Therefore, the final conclusion is that SelU3 is not found in Varanus Komodoensis. To confirm that, we checked if this protein is found in the chicken, but the protein does not appear either. From an evolutionary point of view, it can be said that the ancestor of the the species tested (komodo dragon, lizard and chicken) did not acquire the protein or had already lost it. However, the human ancestor did acquire SelU3.
Thioredoxin Reductases (TR)
Thioredoxin reductases (TRs) are oxidoreductases that, together with thioredoxin (Trx), comprise the major disulfide reduction system of the cell. In mammalian cells, there are three TR isozymes, all of which are Sec-containing proteins. These proteins contain a Sec residue in the COOH-terminal penultimate position12.
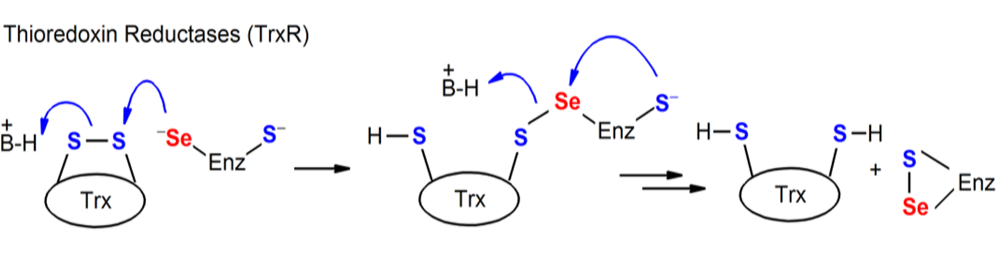
Figure. Chemical processes mediated by Sec-containing enzymes (TR).
This protein is found in both species. In the case of the human, TR1 shows hits distributed in 4 different scaffolds. At the beginning, we were not sure about the scaffold we should use. This is why we run the programme for all of them and then we chose just one. Finally, we chose the hits found in the scaffold SJPD01000048 between positions 5896400-5909749, as it was the one with the T-coffee highest score.
In the case of the lizard, a lot of hits were also found in 4 different scaffolds. First of all, we refuse the scaffold SJPD01000040 because the score of the T-coffee was the lowest (score=656). We finally decided to choose the scaffold SJPD01000048 too because it has a very high score of the T-coffee and a huge gap was found in the other two scaffolds.
Therefore, regarding the results obtained in the scaffold chosen, the predicted protein from the human has 13 exons in the reverse strand, whereas the one predicted from the lizard has 14 exons, also in the reverse strand. Furthermore, a match between two Selenocysteines is found in both species. In addition, the Seblastian results show the presence of a significant SECIS element in 3’ UTR region (positions 5892054-5891976 for human and 5912471-5912393 for lizard).
Above all, we can conclude that TR1 is a selenoprotein in Varanus komodoensis.
TR2 protein is only present in the human genome. TR2 shows hits distributed in 3 different scaffolds. At the beginning, we were not sure about the scaffold we should use. This is why we run the programme for all of them and then we chose just one. Once the T-coffee was done, we say that the scaffold SJPD01000007 was the one that showed a better score (998). However, the other scaffolds also had very good scores, but there was a huge gap at the beginning, meaning that the initial part of the protein was missing.
Therefore, regarding the results obtained with the scaffold SJPD01000007, 7 hits were found. The protein predicted has 3 exons. The score of the T-coffee is very good but there is a gap at the beginning, which means that the initial part of the protein is missing. Moreover, any Selenocysteine was found in the predicted protein and the Seblastian did not predict any significant SECIS element.
Above all, we can say that TR2 is present in Varanus komodoensis but it is not a selenoprotein.
This protein is present in the human genome and in the lizard genome. The blast results show hits distributed in 3 different scaffolds in the case of human and in 4 different scaffolds in the case of the lizard. In both species, we finally decided to choose the scaffold SJPD01000007. We made this choice regarding the phylogenetic tree, as all the scaffolds presented a huge gap at the beginning, which could be due to an incorrect prediction of the structure of the gene in exonerate. However, it has to be said that the lizard TR3 protein has a smaller gap at the beginning than the human TR3 protein, so it could be considered a better alignment. Nevertheless, the number of lacking amino acids is smaller because the lizard protein is smaller as well, which could be due to a shortening of the protein after the split of reptiles.
Therefore, regarding the results in the scaffold chosen, the hits were obtained between positions 20925190-20939399 and 20905682-20939417 in human and lizard respectively. Regarding the protein predicted from the human, it has 15 exons, whereas the protein predicted from the lizard has 16 exons, both in the forward strand. In both cases, a selenocysteine is aligned with another selenocysteine. Furthermore, the Seblastian search shows a significant SECIS element in the 3’ UTR region (between positions 20941512-20941595 in human and 20901512-20901595 in lizard).
For all these reasons, we can say that TR3 is a selenoprotein in Varanus komodoensis.
We tried to perform a phylogenetic tree in order to confirm that the scaffold chosen in each case was the correct, but the tool used did not find any relevant alignment site, which usually means the alignment is not reliable. However, we know that, as these proteins belong to the same family, they should present homology. This is why we tried again the phylogenetic workflow without GBlocks, which eliminates poorly aligned positions. Therefore, although we have to be aware that this phylogenetic tree may not be 100% reliable, we can have an idea of the scaffolds that are more similar to the human query protein.
Regarding the tree, we can see that the scaffolds chosen are correct except in TR2. We chose the scaffold SJPD01000007 but maybe the scaffold SJPD01000047 presents more homology with the query protein.
Selenoprotein machinery
Sec-specific eukaryotic elongation factor(eEFsec)
This protein works as a Sec-specific eukaryotic elongation factor, recruiting tRNA[Ser]Sec and inserting the Selenocysteine as the UGA codon residue into de novo translated protein. This specificity is thought to be directed by the presence of SECIS elements in the 3’UTR region of the protein8.
All the genes of eEFSec from both reference species, one from human and three from lizard, were found in the SJPD01000007.1 scaffold.
When blasting, one of the lizard protein aligned in the reverse strand (18630044-18649127) similarly to the human one (18589990-18701233). The latest was predicted 6 exons in the reverse strand, whereas in the protein from lizard 3 exons were found. For each of the other two lizard proteins, 18580056-1859700 and 18648900-18649127, only one exon was annotated. Regarding the T-coffee results, a better global alignment is obtained from the lizard prediction, explained by the evolutionary proximity. While from the human prediction, less correspondence is observed, as an initial gap of 74 bp and a final one of 63bp are spotted.
To clarify the misleading data, the chicken’s eEFsec was considered, resulting with an alignment comprising all the exons annotated previously. With all that, and considering the proximity, almost overlapping, of the alignment and the alternation of exons from the different reference proteins and species, we can say that probably the lizard protein’s annotation from Selenodb is incomplete. However, it is confirmed that the eukaryotic eEFsec protein is conserved in Varanus komodoensis’ proteome.
In this case, no characteristic features from selenoproteins could be predicted, which makes sense being eEFSec a protein of selenoproteins machinery, so it is not a selenoprotein, confirming our assumptions.
phosphoseryl-tRNA kinase (PSTK)
PTSK (phosphoseryl-tRNA kinase) phosphorylates Ser-tRNA[Ser]Sec to produce the phosphorylated intermediate PSer-tRNA[Ser]Sec, serving as a substrate for SecS.
This protein is not found in the human genome, only found in lizard’s. When running the blast, 4 hits are found in the scaffold SJPD01000060.1 between positions 674799-683674. Also, from the global alignment obtained through T-coffee a good alignment can be seen. The predicted protein has 6 exons in the forward strand.
Regarding the selenoproteins characteristics, it does not contain a the Selenocysteine as we were expecting because we know that this protein is part of the machinery and not a selenoprotein itself. Although the Seblastian predicts one SECIS element it is located more than 10.000 base pairs away from our gene, so we consider that it is not valid.
Regarding the results, we can conclude that PSTK is present in Varanus komodoensis but it is not a selenoprotein.
SECIS binding protein 2 (SBP2)
SECIS binding protein 2 (SBP2) is one of the two trans-acting factors are required for efficient recoding of UGA as Sec in eukaryotes. SBP2 is stably associated with ribosomes and contains a binding domain that is known to bind SECIS elements with high affinity and specificity 8.
This protein is present in both human and lizard. When running the blast, different hits are found in two different scaffolds in both species. We finally decided to choose SJPD1000041.1 as it was the one that presented better alignment in the T-coffee.
Regarding the results obtained in the scaffold chosen, the hits were obtained between positions 7176579-7175318 and 7191828- 7171816 for human and lizard respectively. The protein predicted from the human has 6 exons, whereas the protein predicted from the lizard has 16 exons, both in the reverse strand. Although both scores of the T-coffee are the same (997), the protein predicted from the human presents a big gap at the beginning, which means that part of the protein is missing. Though, the protein predicted from the lizard does not present any gap, which makes sense since they are closely related species.
Analyzing the selenoprotein characteristics, we do not find any alignment between two selenocysteines and no SECIS element is found in the 3’ UTR region. However, we can see the alignment between cysteines. In the case of the lizard, SBP2 is a cysteine-containing homolog or a selenium machinery protein (both things are considered in SelenoDB 2). Nevertheless, we can not determine if one of those cysteines was a selenocysteine in the past or not. Moreover, any SECIS was found in the 3’ UTR region and Seblastian did not predict any known selenoprotein.
Therefore, taking into account all the results, we can say that SBP2 is present in Varanus komodoensis but it is not a selenoprotein.
Sec synthase (SecS)
The conversion of the serine moiety on tRNA[Ser]Sec to selenocysteyl-tRNA[Ser]Sec is catalyzed by Sec synthase (SecS), which incorporates selenophosphate, the active form of Se, into the amino acid backbone and forms Sec-tRNA 8.
This protein is only present in the lizard genome. When running the blast, 10 hits were obtained in the scaffold SJPD01000012.1 between positions 13832197-13801608. The predicted protein has 11 exons in the reverse strand. Regarding the T-coffee results, a perfect alignment can be seen and it starts with a methionine, so it is probably a real protein. The protein predicted does not contain a Selenocysteine as expected, because it is part of the machinery and it is not a selenoprotein itself. Moreover, not any valid SECIS element was found using Seblastian and also any selenoprotein was predicted in the sequence.
Therefore, we can confirm that Varanus komodoensis has SecS and it is not a selenoprotein.
SELENOPHOSPHATE SYNTHASE (SPS or SPHS)
This family of Selenoproteins consists of two proteins involved in selenophosphate synthesis with high homology to SelD from E. coli. Recently, new functions have been discovered, distinguishing SPS2 as a de novo selenophosphate synthase, while SPS1 may have a possible role in Sec recycling through a selenium rescue system, interacting with Sec Lyase8.
SPS1 can be found in the two reference species. When running the blast, two scaffold aligned for this protein: SJPD01000051.1 and SJPD01000016.1. The latest was chosen, considering homology arguments, as indicated in the phylogenetic tree. The multiple hits for both proteins were in the reverse strand from 15068382-15092907 positions. The predicted protein had 8 exons in the reverse strand. Plus, with a methionine in the start point, a score of 1000 and a total alignment, except from three mismatches, the T-coffee results indicate the existence of this protein in its entire form.
As it was expected, no characteristic features from selenoproteins were found, confirming our assumptions.
Overall, the data indicates the existence of SPS1 in Varanus komodoensis’ proteome.
This protein is found in both the reference species. Similarly to the protein from above, the same two scaffolds aligned with it. The phylogenetically closest one (SJPD01000051.1) was used to perform the analysis. The human gene aligned in the reverse strand from the position 3708299 to3713123, around 1700bp less than the lizard gene (3708260-3713129). After the prediction, the exonerate of the human protein was also incomplete, 7 exons were annotated, compared to the 8 exons found in the lizard. Interestingly, as the missing exon (3714627-3714927) is the one containing the expected Selenocysteine.
The evolutionary distance becomes more evident when considering the T-coffee results, the human prediction showed a poor global alignment of a 991 score, with an initial gap of 113bp, in which the methionine and the Sec residues are skipped. Whereas the lizard prediction the alignment, of 998 score, was complete.
Seblastian software predicts the same Selenocysteine as in our prediction, plus one element of A grade in the corresponding-strand 3’UTR region at a reasonable distance (3707840-3707915).
Considering that, we can extract that the selenoprotein SPS2 is found in the Varanus komodoensis’ proteome.
A phylogenetic tree of the protein family was constructed in order to distinguish the corresponding scaffold of each Selenophosphate synthase, as they presented a strong sequence similarity from its familiar origin. From its proximity, SJPD01000016 scaffold is considered the region comprising SPS1 protein in our specie’s genome and SJPD01000051 for SPS2 protein.
(SECp43)
SECp43 interacts with the tRNA[Ser]Sec forming a complex. It has a nuclear localization and it may work as a chaperone for SLA and Sec-tRNA[Ser]Sec, being linked to the regulation of the synthesis of selenoproteins through methylation of tRNA[Ser]Sec and the intracellular distribution of SLA 8.
This protein is only present in the case of the lizard. When we run the blast, only one significant scaffold is found (SJPD01000036.1) and the hits obtained are located between positions 4926809-4935614. The predicted protein has 4 exons in the forward strand. T-coffee shows a good identity, although it presents a gap at the beginning, maybe because exonerate has not correctly predicted the gene structure and, therefore, these firsts amino acids are missing (including the methionine). As expected, no alignment between two selenocysteines was found and the Seblastian did not predict any SECIS element and also no selenoproteins were predicted.
For all these reasons, we can confirm that Varanus komodoensis has SECp43 and it is not a selenoprotein.