Discussion
DIO (Iodothyronine Deiodinases) Family
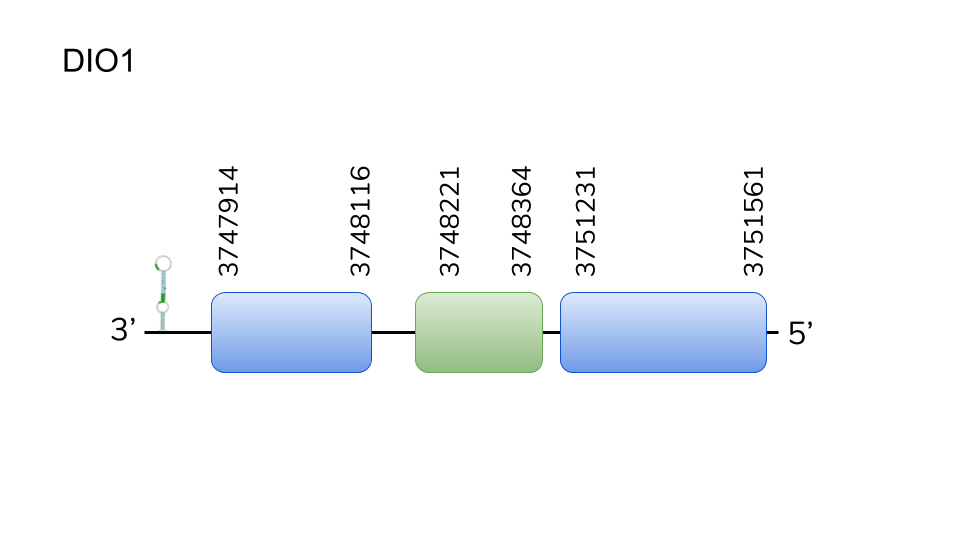
DIO1:Several hits in three scaffolds were retrieved from BLAST for DIO1, being the hit in contig UELW000089.1 the best one. T-coffee prediction of the alignment between the best hit-predicted and reference proteins yielded a coding sequence for DIO1 in the Danionella dracula genome. The predicted gene is located at 3747914 - 3751561, and is composed of 3647 nucleotides, comprising 3 exons, as predicted by exonerate. Seblastian prediction yielded a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian predicted a protein in the same scaffold as Danionella dracula’s DIO1. Therefore, we concluded DIO1 can be found as a selenoprotein in the genome of Danionella dracula.
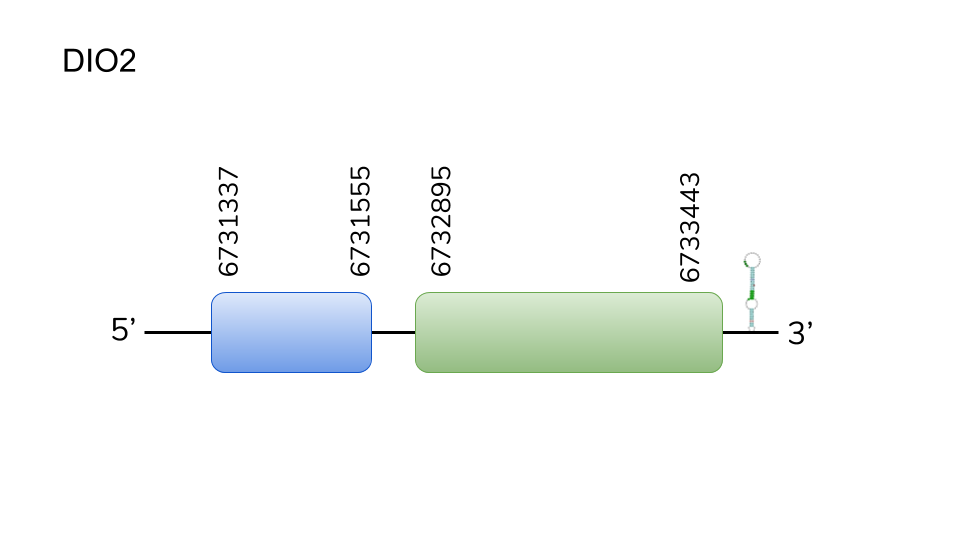
DIO2: In a similar manner as the previous case, BLAST search for the sequence of Danio rerio’s DIO2 protein in the genome of Danionella dracula yielded hits in three different scaffolds. The best hit was located in the scaffold UELW01000223.1, and the predicted protein, analysed using exonerate and T-coffee, was located in positions 6731337 - 6733443 of the genome of D.dracula, has a length of 2106 nucleotides, which include 2 exons.
Analysing the scaffold using Seblastian resulted in the prediction DIO2 a as a selenoprotein. A SECIS element was predicted 3’ from the predicted gene. Taking this results into account, it can be concluded that DIO2 is a selenoprotein in Danionella dracula.
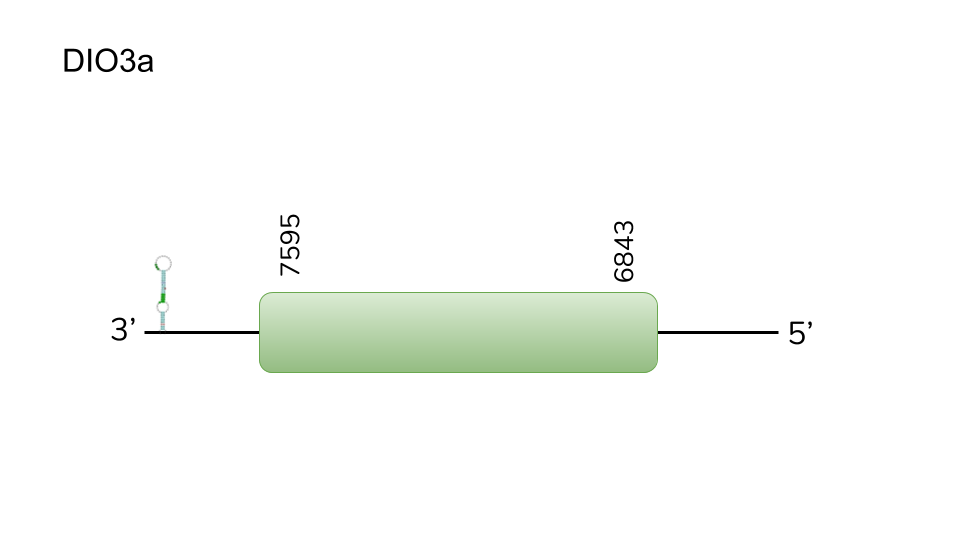
DIO3a: We found one hit in scaffold UELW01000069.1, The BLAST hit for scaffold UELW01000069.1 resulted in the best score searching Danio rerio’s DIO3a, several other hits were found in other scaffolds, with lower scores. The genomic position of the hit predicted by exonerate in the scaffold is 6843 - 7595, having a length of 752 nucleotides in the genome of Danionella dracula. T-coffee prediction of the alignment between the predicted protein and Danio rerio’s DIO3a yielded a high-scoring alignment. The scaffold was analysed using Seblastian, yielding the prediction of a selenoprotein in the genome. A SECIS element was predicted 3’ of the predicted gene, which led us to the conclusion that DIO3a is a selenoprotein in Danionella dracula.
DIO3b:The analysis of Danio rerio’s DIO3b protein yielded the same positive hits in the same scaffolds as DIO3a. Analysing the best hit, it was found that, in fact, Danio rerio’s DIO3b is equivalent to DIO3a in the genome of Danionella dracula, suggesting that this species has lost the protein during its evolutionary timeline.
eEFsec:Hits for eEFsec were found using BLAST in scaffold UELW01000234.1, 7 hits were selected. The protein is located in the genome in positions 4166591 - 4176749, with a length of 10158 nucleotides, in the positive strand. 7 exons were identified. The predicted sequence showed a high-scoring alignment with the reference protein. Seblastian predicted a SECIS element 3’-UTR. However, it could not predict a sequence, thus demonstrating that we can consider the presence of eEFsec as a Cys-containing homolog in the selenoprotein machinery of Danionella dracula.

GPx Family (Glutathione peroxidases):
While analyzing the results of this protein family, we found that there was a relatively big number of hits for each protein, distributed along several scaffolds that were common to several proteins of the family. After the analysis we concluded that these spurious hits corresponded to segments of queries from other proteins of the same family, and we were able to discard them.
GPx1a: Two hits were selected in scaffold UELW01000234.1. The protein is located between 3560389-3561798 positions in the positive strand, with a length of 1409 nucleotides. Two exons were identified, the first of them containing the Se-Cys. The predicted sequence showed an excellent alignment with the query in the T-Coffee analysis, being the selenocysteine among the aligned amino acids. Seblastian also predicted a SECIS element in 3-’UTR. With the data collected we consider GPx1a to be present in Danionella dracula.
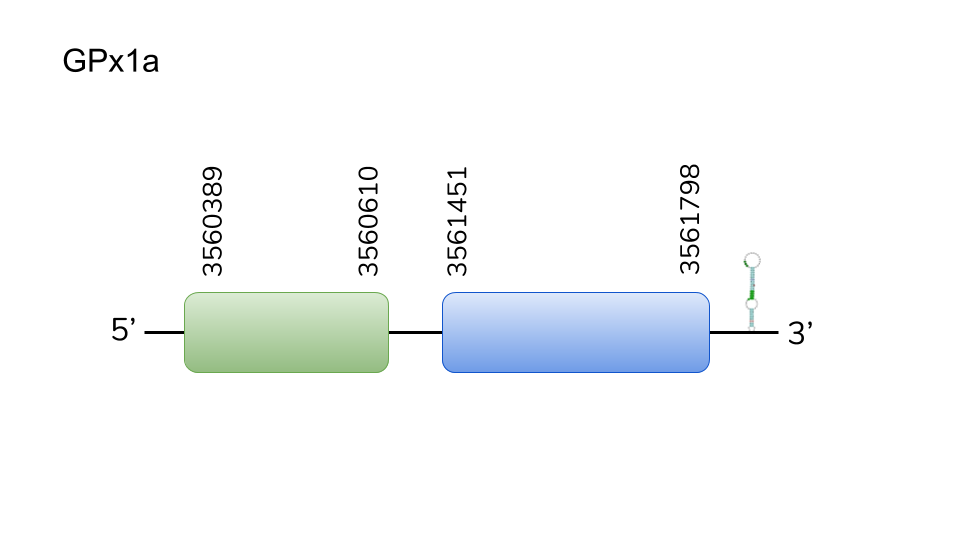
GPx1b: We found two hits in scaffold UELW01000012.1. Predicted GPx1b is located 8872075-8873398 positions in the reverse chain with a length of 1323 nucleotides. The protein is composed by two exons. T-Coffee alignment with Danio rerio’s query showed a great homology between the two species’ protein, including selenocysteine position. A SECIS element was predicted in 3’-UTR. Therefore, we concluded the presence of this protein in Danionella dracula’s selenoproteome.
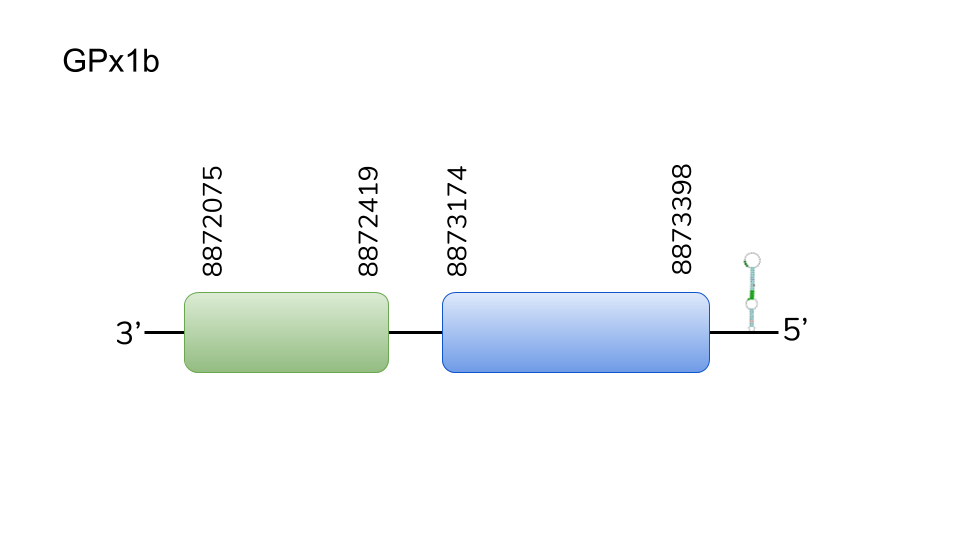
GPx2:Two hits were found in scaffold UELW01000241.1. Located in 27286-29547 positions in the negative strand with a length of 2261 nucleotides . A selenocysteine residue was found in the first of two exons. T-Coffee revealed a solid homology with Danio rerio’s query, including Se-Cys. Moreover a SECIS structure was predicted in 3’-UTR. This evidence lead us to expect GPx2 in Danionella dracula’s selenoproteome.
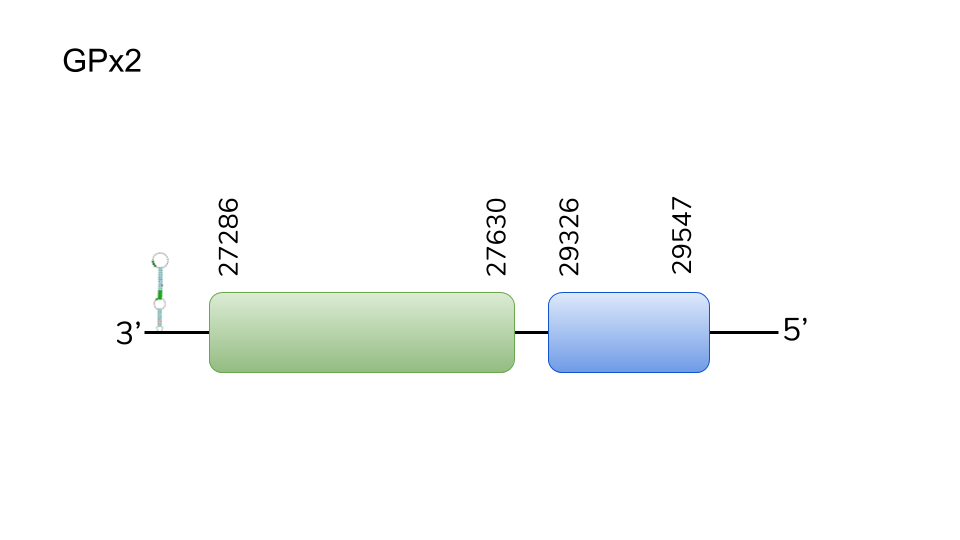
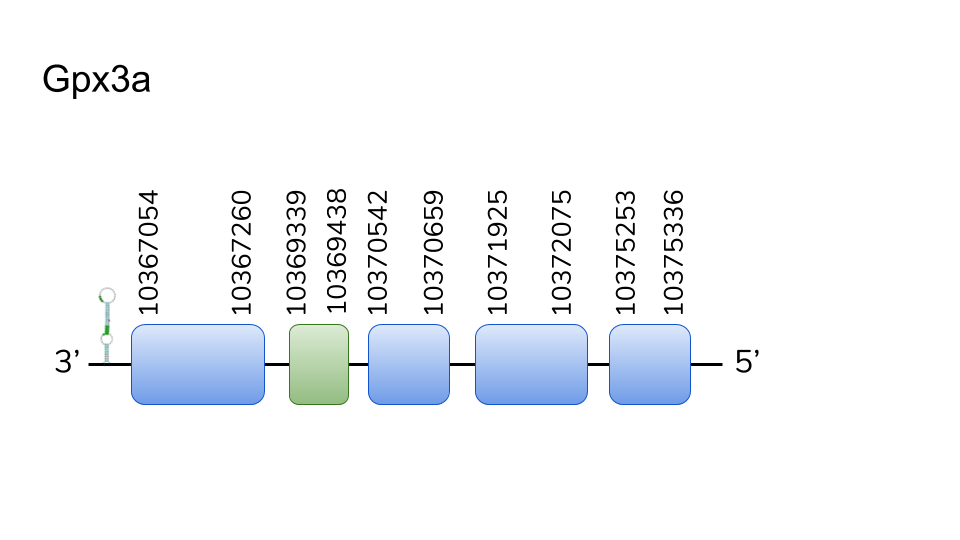
GPx3a: four hits were found in scaffold UELW01000001.1. Located in 10367054-10375336 positions of the negative strand with a length of 8282 nucleotides. Five exons were predicted for this protein, the second containing a selenocysteine. T-Coffee revealed a great conservation between the two species, including selenocysteine alignment. Two SECIS elements were predicted for 3’-UTR of GPx3a. All in all, the evidence provide allows us to consider GPx3a as a Danionella dracula selenoprotein.
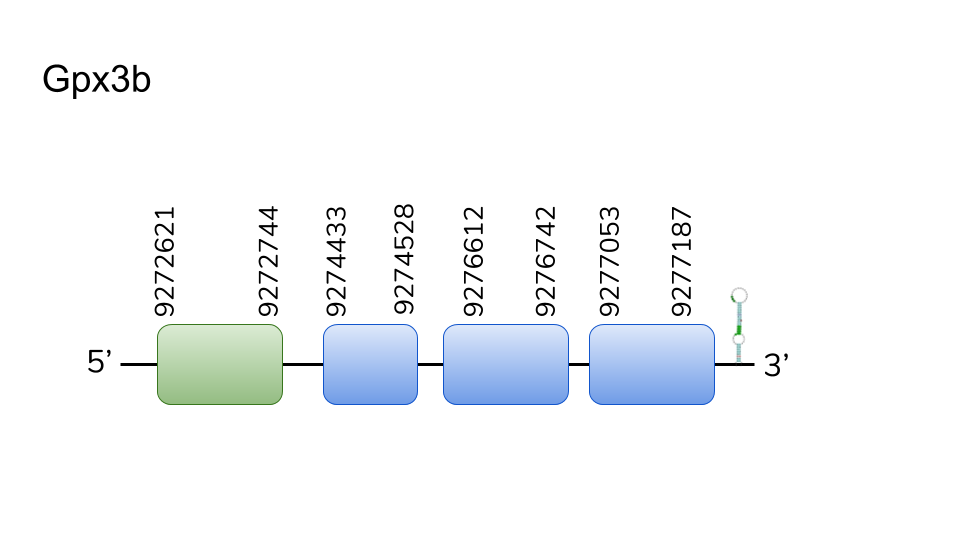
GPx3b: Three hits were found in scaffold UELW01000222.1. Located in 9272621-9277187 positions of the positive chain with a length of 4566 nucleotides. GPx3b is composed by 4 exons, being the first the container of a selenocysteine. T-Coffee results were positive overall, including Sec alignment. However, Danionella dracula’s GPx3b seems to lack the last residues of C-terminus with regard to the query. Even though no selenoprotein could be predicted by Seblastian, the presence of a SECIS element in 3’-UTR as well as the exonerate and T-Coffee results makes us expect the presence of GPx3b in Danionella dracula’s selenoproteome.
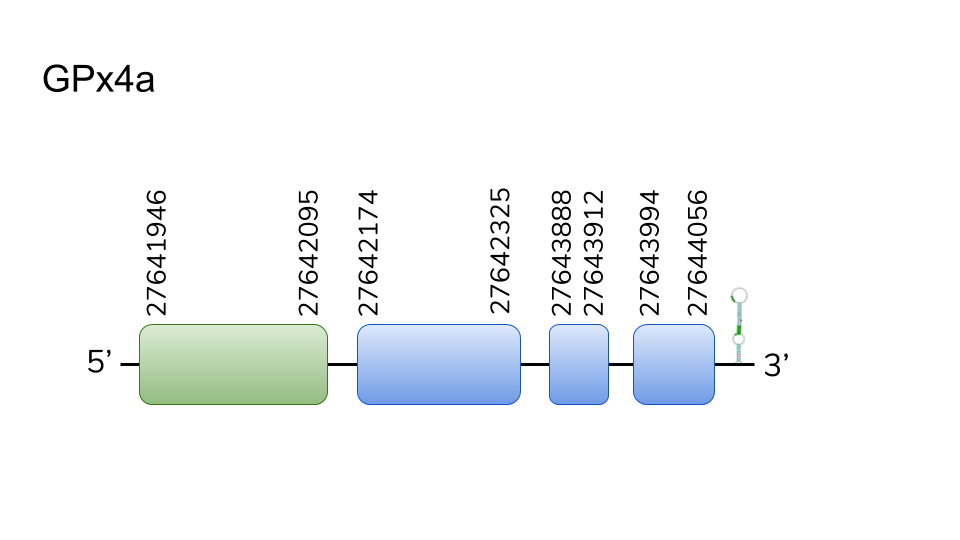
GPX4a:We found two hits in scaffold UELW01000778.1. Located in 27641946-27644056 positions in the positive strand with a length of 2110 nucleotides. 4 different exons were described for this protein with a Sec present in the first one. T-Coffe alignment was excellent with no STOP codons before C-Terminus and Sec alignment for the query and the predicted protein. A SECIS element was also found in 3’-UTR. Taking all the above discussed into account, there is a high likelihood regarding the consideration of GPx4a as one of Danionella dracula’s selenoproteins.
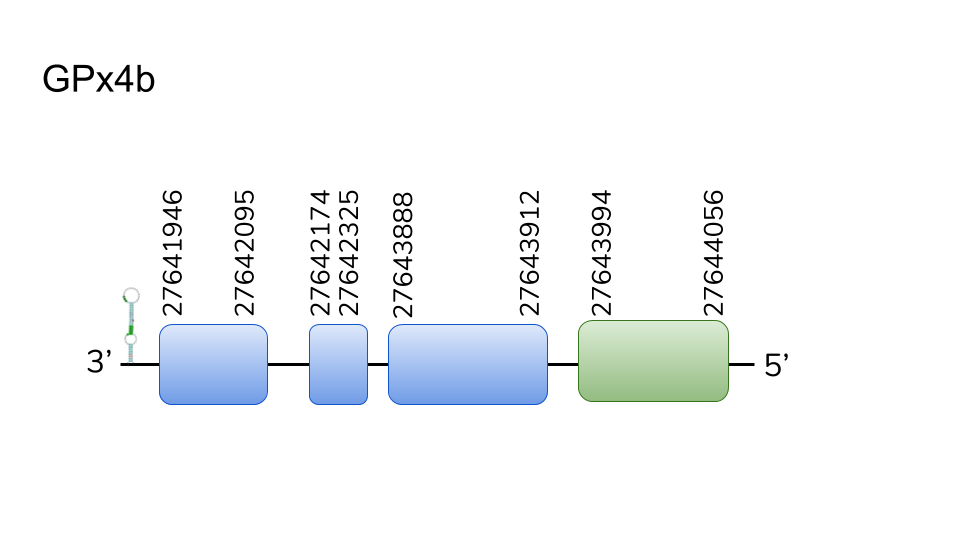
GPx4b: We found 4 hits in scaffold UELW01000721.1. Located in 8795-11608 in the negative strand with a length of 2813 nucleotides. A Sec was found in the fourth of four exons composing this gene. T-Coffe showed almost a perfect alignment of the two sequences, including the Sec residue. Seblastian also predicted a SECIS element in 3’-UTR. We were unable to locate any stop codon before the C-terminus. All of the above incles us to consider GPx4b part of Danionella dracula’s selenoproteome.
GPx7: We were unable to find correct hits corresponding to this protein. Our best hit shared a great homology with GPx8 prediction lacking the first sequence of this last. Moreover the predicted sequence for GPx7 contained a stop codon before the C-terminus.
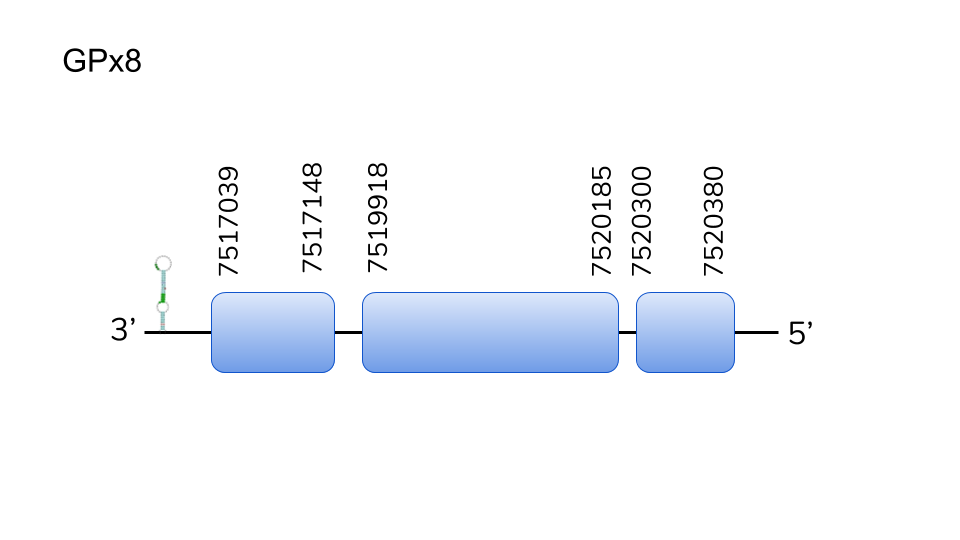
GPx8:Two hits were found in scaffold UELW01000666.1. Located in 7517039-7520380 position of the reverse chain with a length of 3341 nucleotides. It is composed of 3 exons and none of them contains a Sec. This confirms our suspect of being a Cys-homologue, as it was for our query. T-Coffee results confirmed a high homology between the query and predicted protein. Despite no having a Sec residue, a SECIS element was predicted in 3’-UTR. This data highly suggests to include GPx8 as one of Danionella dracula’s selenoproteins.

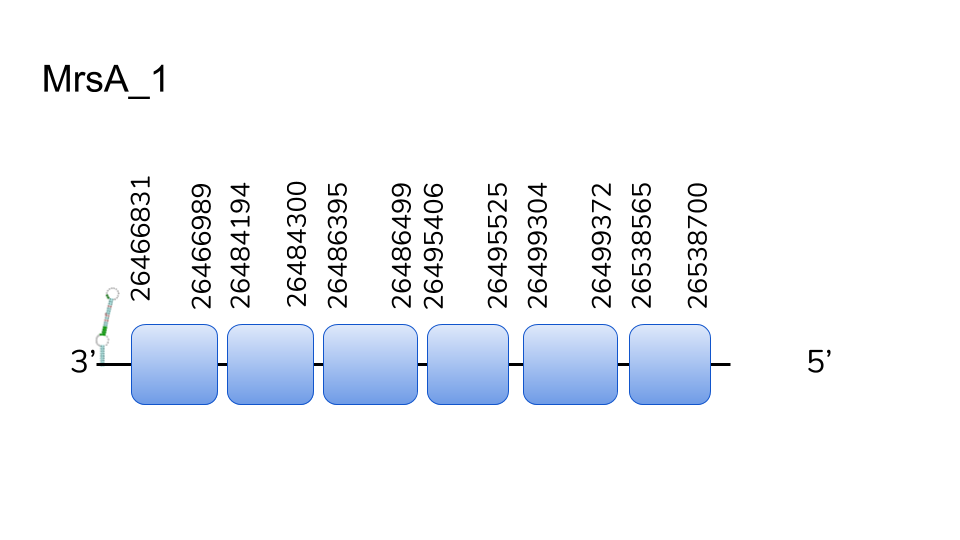
MsrA:BLAST search for Danio rerio’s MsrA protein yielded relevant hits across two scaffolds; UELW01000001.1 and UELW01000223.1. Analysis of both proteins using exonerate and T-Coffee resulted in high-scoring alignments of the predicted to the reference protein in both cases.
The hit in scaffold UELW01000223.1 ranges from positions 418314 to 434307 of the D.dracula genome, spanning 15993 nucleotides. Analyses performed using Seblastian yielded proteins, which drove to the hypothesis that MsrA has undergone a duplication in the genome of Danionella dracula. Furthermore, prediction of SECIS elements both 3’ of the predicted gene we considered that MrsA exists as a selenoprotein in the genome of D.dracula.
MsrB (Methionine r-sulfoxide Reductase) Family
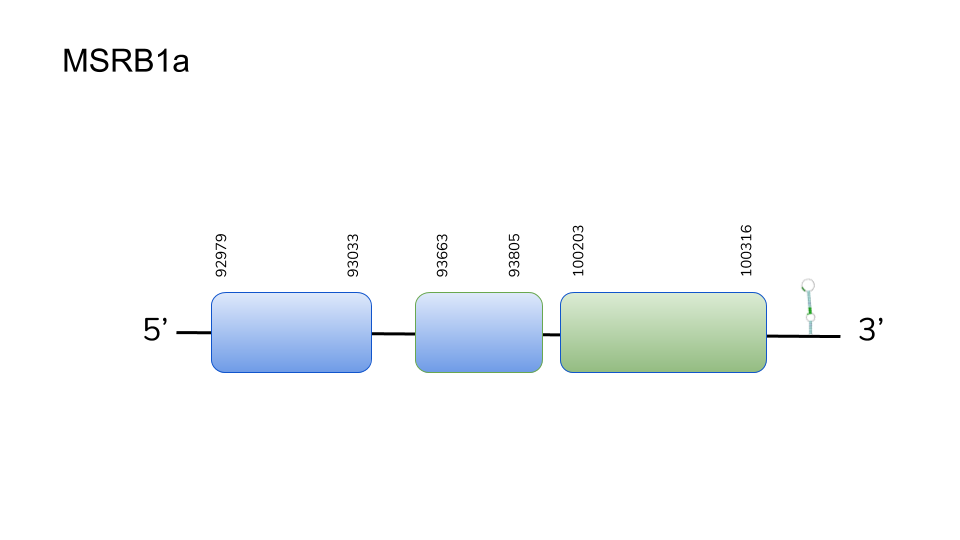
MSRB1a: BLAST search for Danio rerio’s MSRB1a yielded positive results for two scaffolds in the genome of D. dracula; UELW01000644.1 and UELW01000800.1. Further analysis using exonerate and T-Coffee resulted in scaffold UELW01000644.1 containing the best predicted protein, as it yielded the best-scoring alignment, including Sec alignment (on the third exon), in the positive strand.
The predicted MSRB1a gene is found in scaffold UELW01000644.1 can be found in positions 92979 to 100316 of the genome of Danionella dracula, and spans 3 exons.
Seblastian analysis for the best hit and alignment resulted in the prediction of a selenoprotein in the genomic region of choice. Basing on the prediction of a SECIS element 3’ from the gene, we can conclude that MSRB1a is a selenoprotein in Danionella dracula.
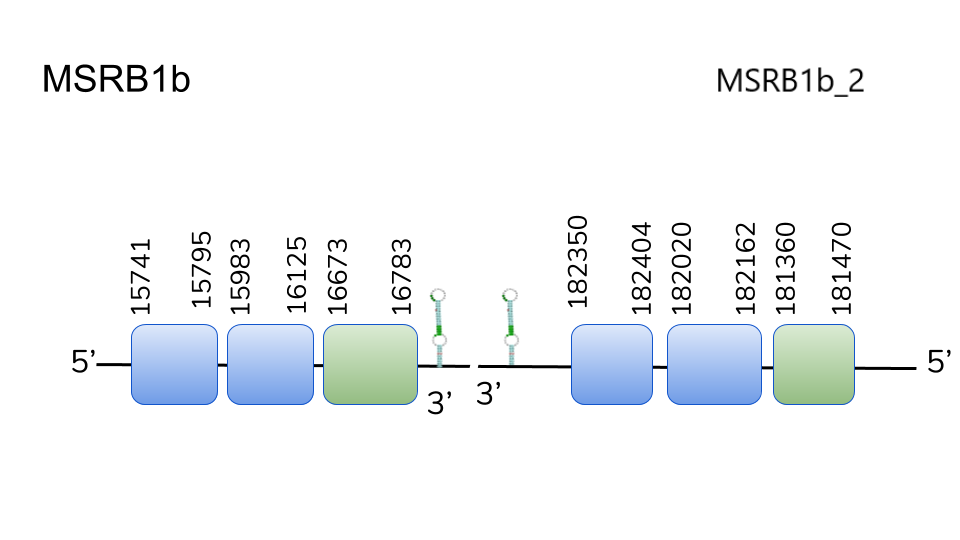
MSRB1b and MSRB1b_2: Like in the previous case, hits were found in the same scaffolds (UELW01000644.1 and UELW01000800.1) using BLAST. Opposite to MSRB1a, it was scaffold UELW01000800.1 which yielded the best-scoring alignments (including Sec alignments) and hits, and thus the proteins were predicted.
MSRB1b predicted genes in Danionella dracula were predicted in positions 15741 to 16783 (positive strand) and 182350 to 181470 (negative strand) of the genome, spanning 3 exons each. These findings allows us to conclude that MSRB1b went through a duplication process during Danionella dracula’s evolution. Both presented SECIS elements in 3’-UTR leading us to include MSRB1b as a Danionella dracula’s selenoproteins
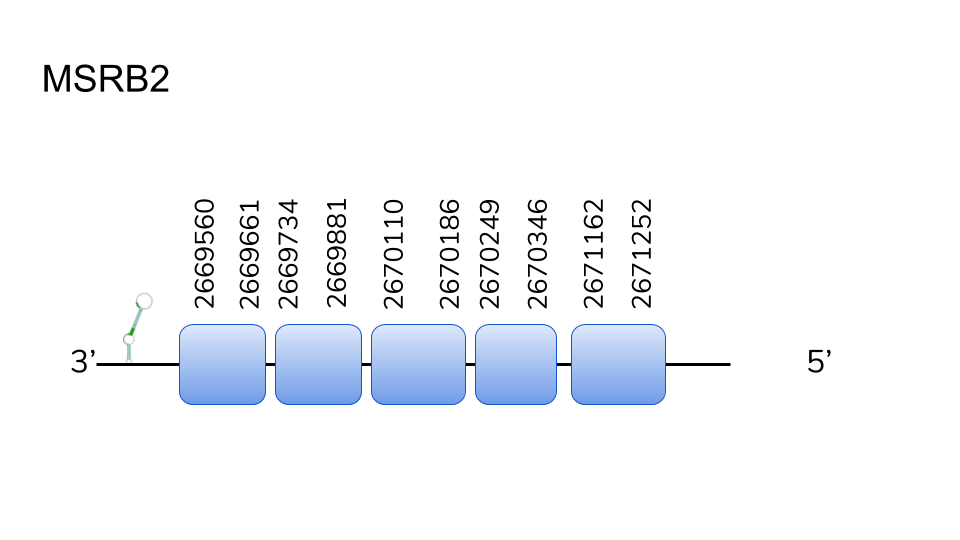
MSRB2: Similarly to the aforementioned case, hits were found in two scaffolds (UELW01000123.1 and UELW01000888.1) were obtained using BLAST. The best-scoring hit and alignment was the result of the analysis of scaffold UELW01000888.1, yielding a gene in positions 2669560 to 2671252 of the genome, which spans 5 exons.
Seblastian analysis of the predicted protein in the scaffold yielded no positive results. However, 2 SECIS elements were predicted using SECISearch3. Thus considering our prediction of MSRB2, taking into account the high-scoring alignments and predictions, to be a Cys-containing homolog present in D.dracula’s selenoprotein machinery.
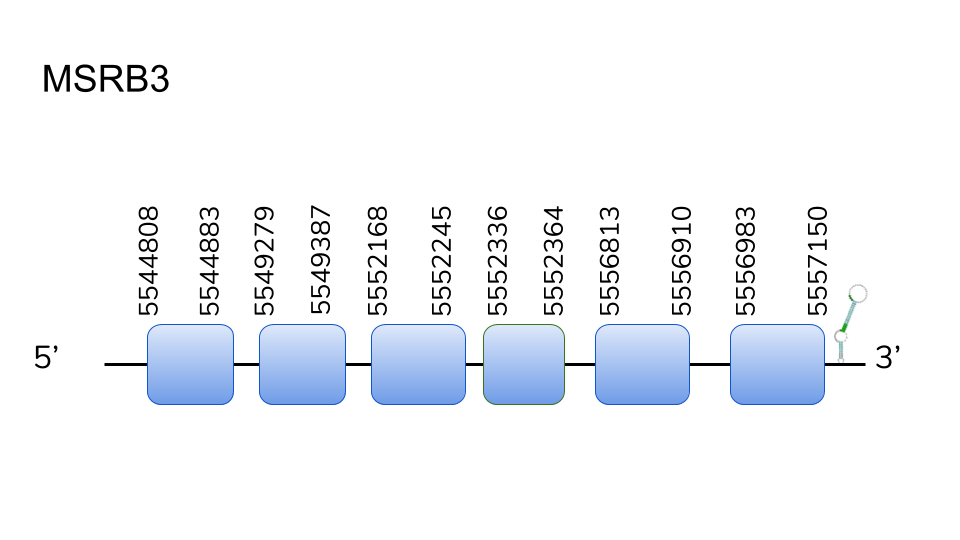
MSRB3:In opposition to the beforementioned case, scaffold UELW01000123.1 yielded the best BLAST hit, meaning that MRSB2 yielded a hit in the MSRB3 scaffold, and vice-versa. T-Coffee alignment yielded high-scoring results, and exonerate determined MSRB3 could possibly be found in positions 5544808 - 5557150 of the contig, containing a total of 6 exons. Like in the previous case, Seblastian could not predict a protein for this genomic region. Therefore, we can consider our prediction of MSRB3, taking into account our alignment and prediction, to be present as a Cys-containing homolog present in D.dracula’s selenoprotein machinery, as a SECIS element was predicted in the 3’-UTR for this region.
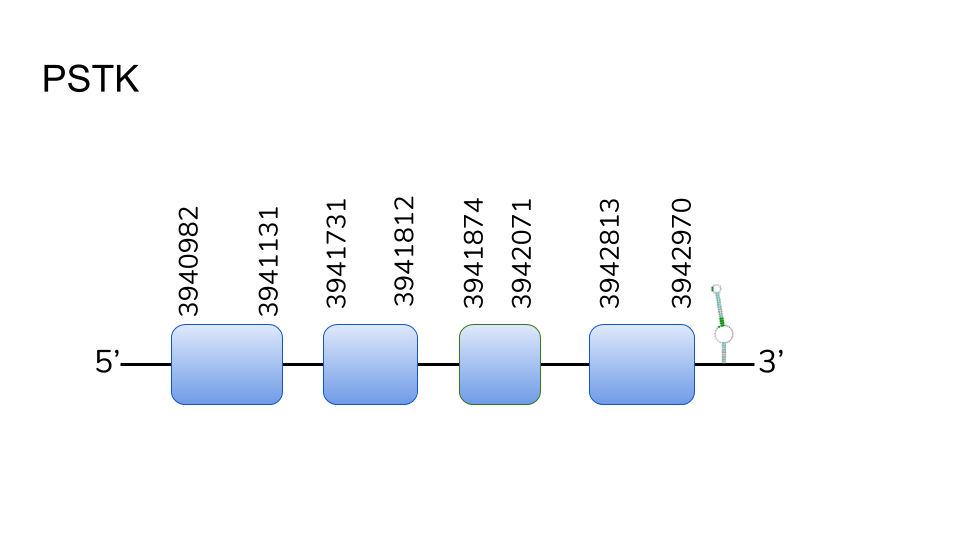
PSTK:All hits for this D.rerio protein were localized in scaffold UELW01000123.1 using BLAST. Using exonerate and T-Coffee a sequence was predicted for the protein in the D.dracula genome. The gene was located in positions 3940982 - 3942970. 4 exons were describes for this protein. Using T-Coffee, the obtained alignment was a high-score one. Additionally, a SECIS element was found in the 3’-UTR. However, no gene prediction yielded from Seblastian analysis. Taking our prediction and high-score T-Coffee alignment into account, as well as scientific references, we can consider PSTK as a Cys-containing homolog present in D.dracula’s selenoprotein machinery.
SBP2: Hits were found using BLAST, in two different scaffolds; UELW01000556.1 and UELW01000552.1. However, after analysis and T-Coffee alignment, identity scores and alignment scores were poor, and not deemed good enough to be considered as copies of a selenoprotein protein in the genome of D.dracula. Furthermore, no SECIS elements were described using SECISearch3, and no prediction was obtained when performing Seblastian searches for any of the scaffolds, thus we considered that D.dracula might have evolutionarily eliminated this protein from its genome.
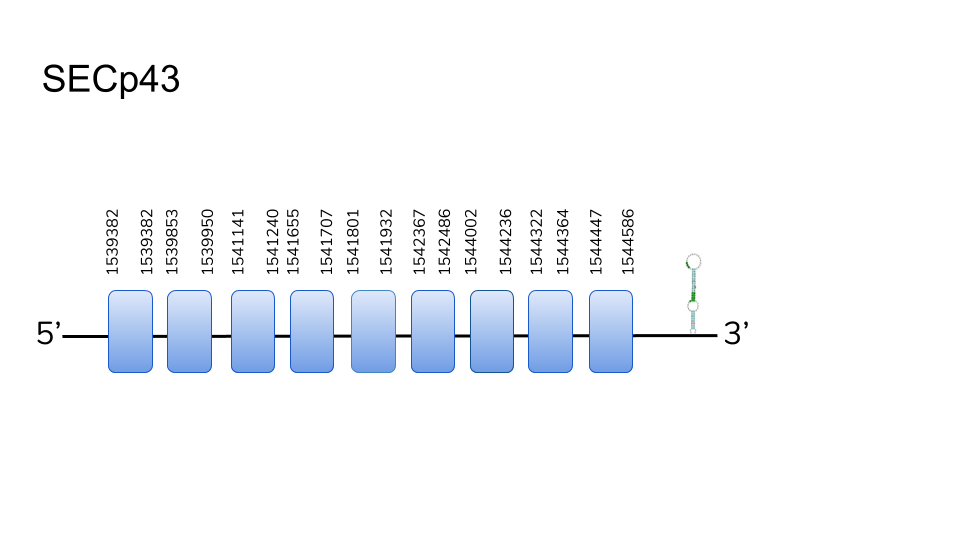
SECp43:5 hits were found in scaffold UELW01000100.1. Located in 1539382-1544586 positions of the positive chain with a length of 5204 nucleotides. 9 exons were found to compound the gene. T-Coffee showed a great alignment between the predicted protein and the query. No stop codons were found before C-Terminus. Although the sequence contains no Sec a SECIS element was described in 3’-UTR. The compiled Data, as well as scientific references allows as to consider SECp43 as Cys-homologue in Danionella dracula’s selenoprotein machinery.
SecS:10 hits were found in scaffold UELW01000053.1. Located in 550525-559495 positions of the positive strand with a length of 8970 nucleotides. SecS posesses 11 exons. A great homology was observed by T-Coffee, only differing in the C-terminus sequence, which seem to be shorter in Danionella dracula. As expected no Sec residue nor SECIS element were described for SecS, leading us to categorize SecS as a probable Cys-homologue with a role in selenoprotein machinery.

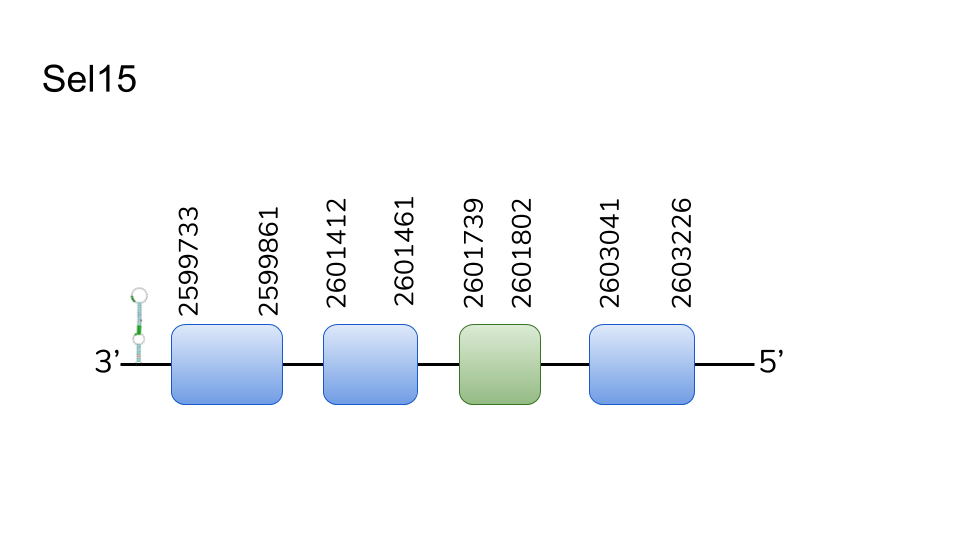
Sel15: We found 3 hits un scaffold UELW01000100.1. Located in 2599733-2603226 position of the negative chain with a length of 3493 nucleotides. 4 exons were predicted with a Sec inside the third. T-Coffee alignment starts quite poorly and contains a stop codon in the sixth residue. However, the rest of the alignment presents a high homology and contains an aligned Sec residue. It is to be considered the lack of a methionine at the start of the sequence. These inconveniences are not present in the Seblastian prediction, in which we find a Met at the start and no stop codons before C-terminus. In addition to this, three SECIS elements were found in 3’-UTR. For the reasons just given, we conceive a high probability of characterizing Sel15 as one of Danionella dracula’s selenoproteins,
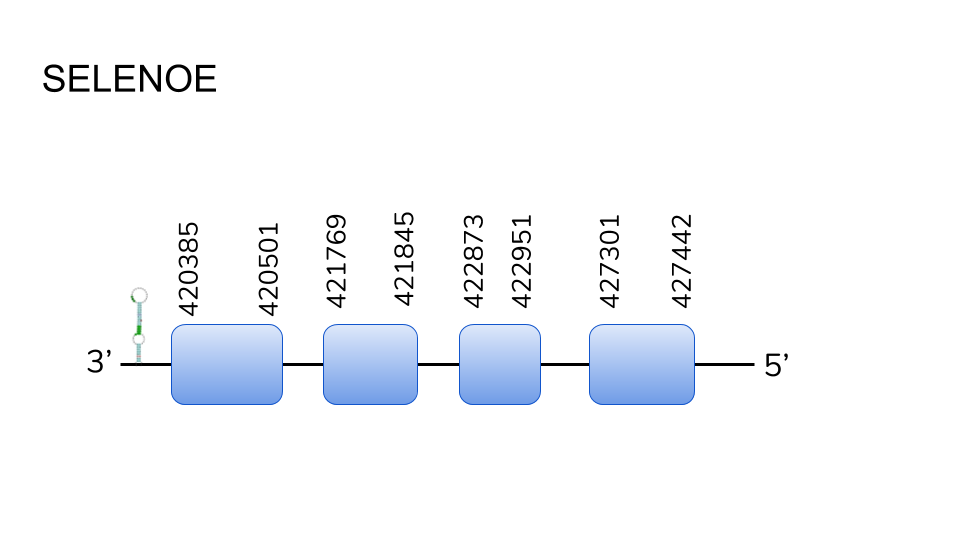
SELENOE: We found 3 hits in scaffold UELW010000766.1, and two more in scaffold UELW01000422.1, which suggests a possible protein duplication. Using Seblastian, we predicted a protein ranging from position 420385 to 427442, with a length of 7057 base pairs comprising 4 exons. No selenocysteine was found in its exons, but 3 SECIS elements were found in the 3’ region of the protein.

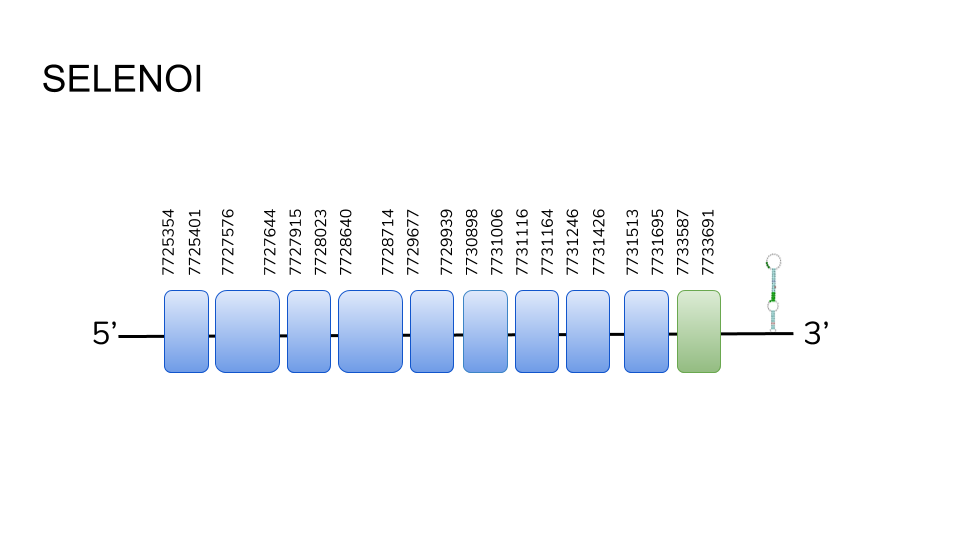
SELENOI: 3 hits for SELENOI were found in scaffold UELW01000766.1, and after analysis through seblastian a protein ranging from position 7225354 to 7733691, with 10 exons along 7057 base pairs was predicted. The protein had one selenocysteine residue in its last exon, and a SECIS element was predicted in the 3’ region.
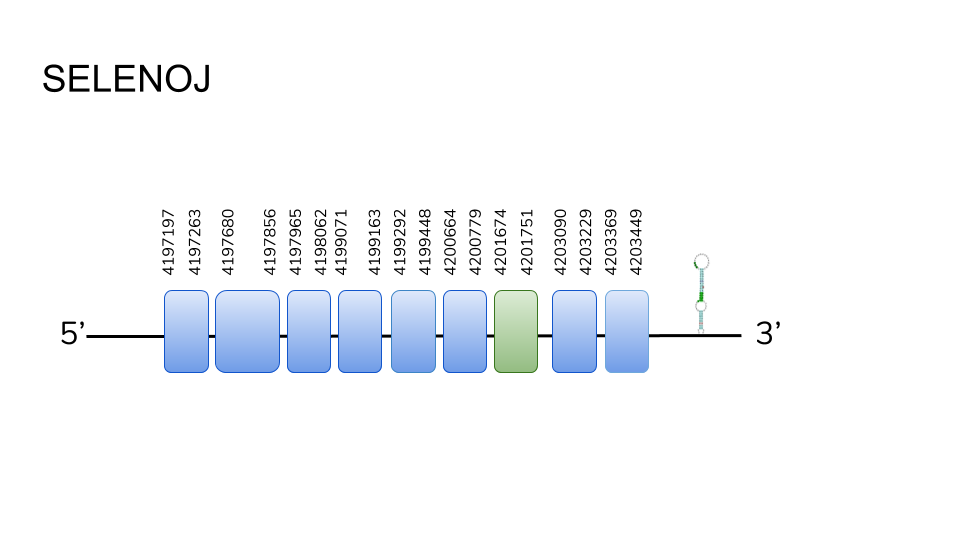
SELENOJ1: Although Seblastian was not able to predict a protein, our analysis found 8 hits in scaffold UELW01000100.1. Our analysis showed a 6252 base pair gene, ranging from position 4197197 to 4203449, comprising 9 exons, with a selenocysteine residue in the seventh, and one SECIS element in the 3’ region.
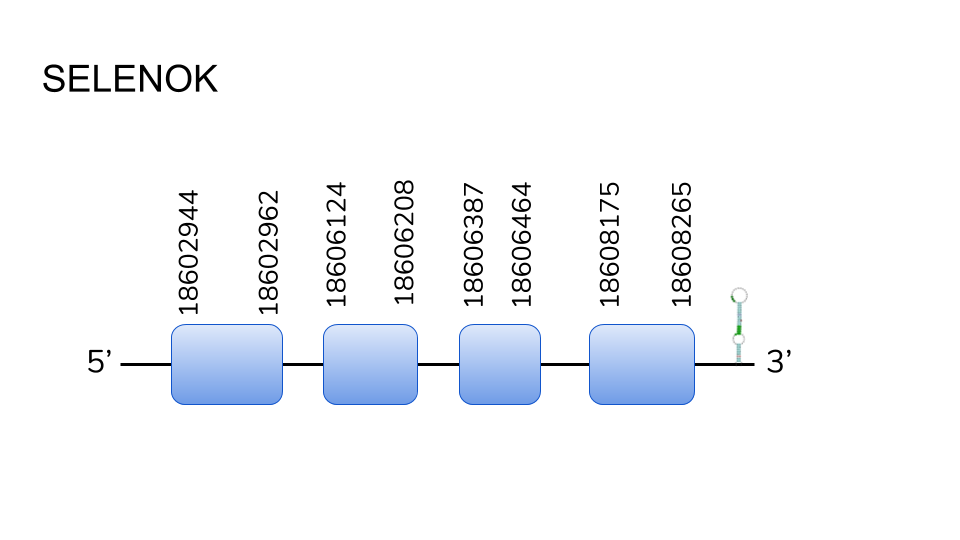
SELENOK: No protein was predicted by Seblastian, but our analysis resulted in 2 hits in scaffold UELW01000334.1, and the prediction of a 4 exon protein ranging from position 18602944 to 18608265, along 5321 base pairs. No selenocysteine residue was seen in any exone, making us conclude that this protein is a cysteine homologue of a selenoprotein ,although two SECIS elements were found in the 3’ region.
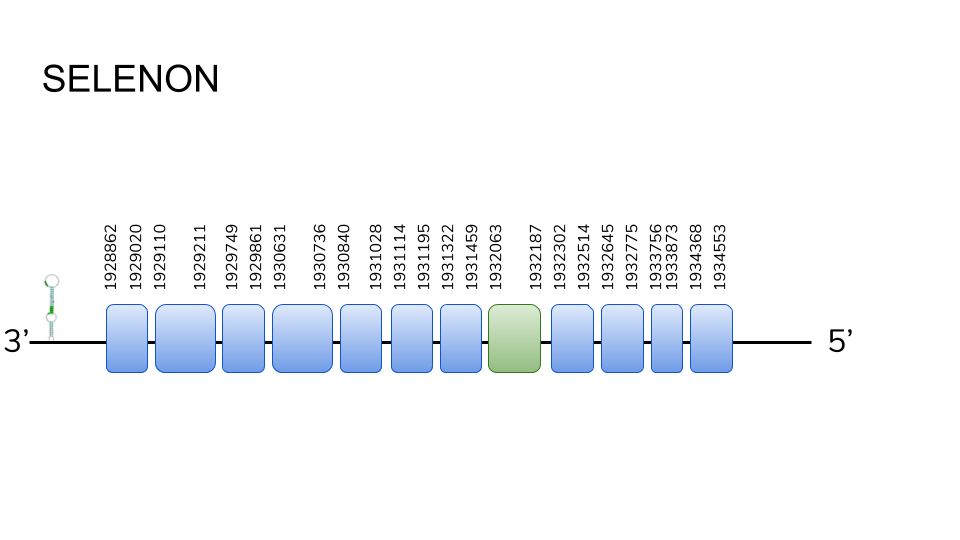
SELENON: 10 hits were found in scaffold UELW01000223.1, and the predicted protein was a 12 exon protein of 591 base pairs along position 1928862 to 1934553. Exon 8 was found containing a selenocysteine residue, and a SECIS element was predicted in the 3’ region.
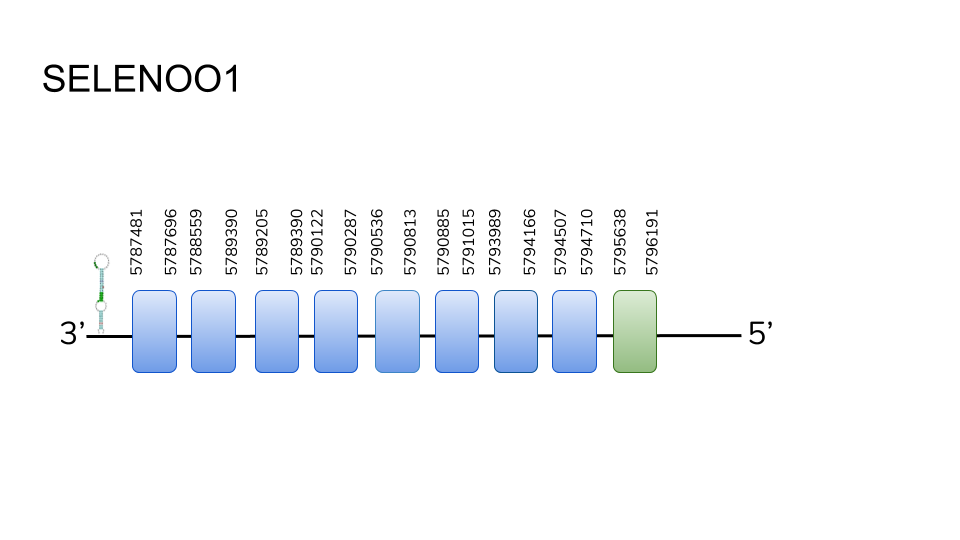
SELENOO1: We found 9 hits in scaffold UELW01000056.1, and were able to predict the gene for a 9 exon protein ranging from position 5787481 to 5796191, with a length of 871 base pairs. The last exon was found to contain a selenocysteine residue, and a SECIS element was predicted in the 3’ region.
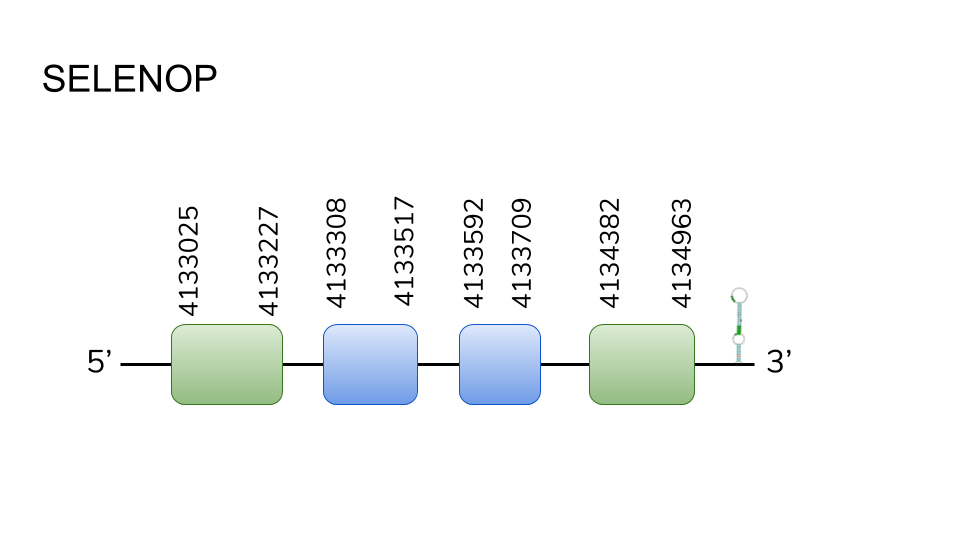
SELENOP: Our analysis found 4 hits in scaffold UELW01000056.1, and 2 hits in scaffold UELW01000056.1. After running the results through t-coffee, only the hits in the first scaffold were found to be suitable enough, and the protein prediction process resulted in a four exon protein that went from position 4133025 to 4134963, with a length of 1938 base pairs. This protein had the particularity of sporting a total of 17 selenocysteine residues, two of those on the first exon and the rest in the fourth exon.
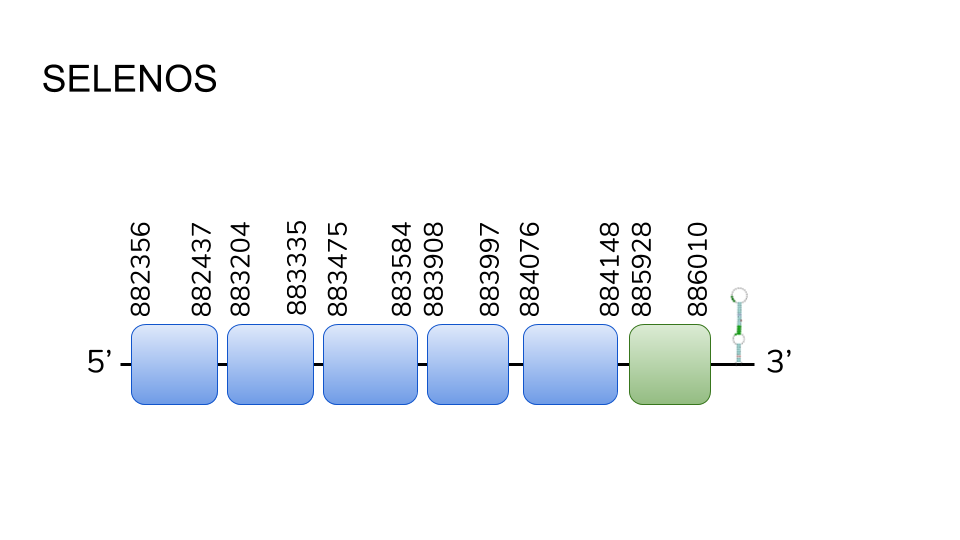
SELENOS: Four hits were found in scaffold UELW01000021.1, and the prediction process resulted in a six exon protein, ranging from position 882356 to 886010 across 3654 base pairs. A selenocysteine residue was found in the last exon, and one SECIS element was found in the 3’ region.
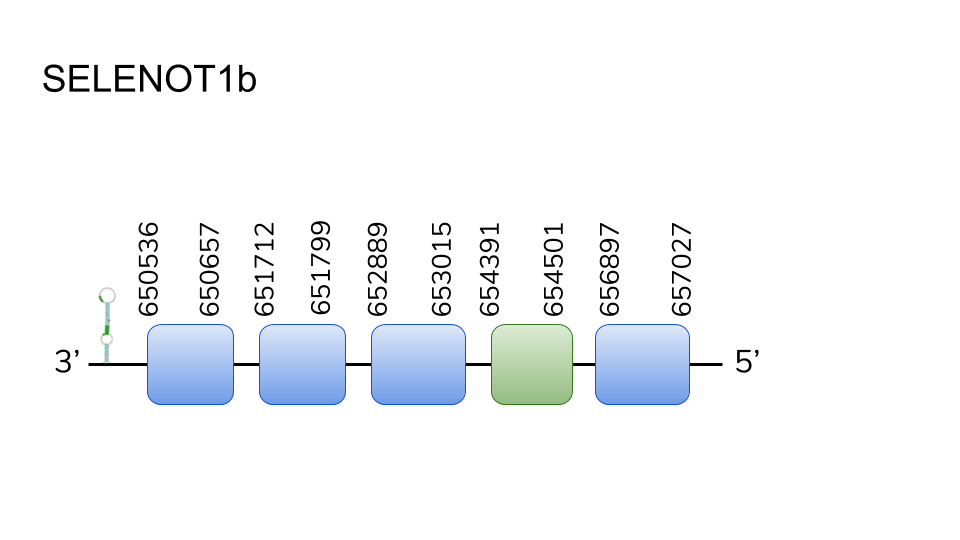
SELENOT1b: Our analysis found 5 hits in scaffold UELW01000178, and 3 hits in scaffold UELW01000788. Analysis through t-coffee deemed the hits in the first scaffold as the most adequate, and protein prediction resulted in a five exon protein that went from position 650536 to 65702 with a length of 6491 base pairs. A selenocysteine residue was found in its fourth exon, and two SECIS elements were found in the 3’ region.
SELENOU1a:Our analysis found several hits along scaffold UELW01000844.1, UELW01000517.1, and UELW1000123.1, but only the 3 hits from UELW01000844.1 were considered good enough after comparison by t-coffee. After predicting the resulting protein, we found a 5 exon protein, from positions 3873461 to 3880391, with a total length of 6930 base pairs and a selenocysteine residue in its second exon. One SECIS element was predicted in the 3’ region

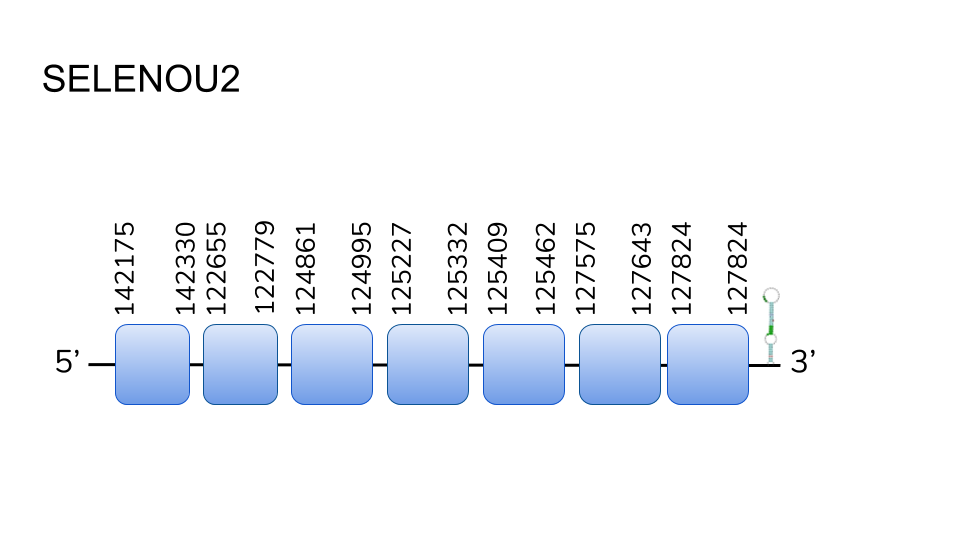
SELENOU2: Our analysis found several hits along scaffold UELW01000844.1, UELW01000517.1, and UELW1000123.1, but only the 3 hits from UELW01000844.1 were considered good enough after comparison by t-coffee. After predicting the resulting protein, we found a 5 exon protein, from positions 3873461 to 3880391, with a total length of 6930 base pairs and a selenocysteine residue in its second exon. One SECIS element was predicted in the 3’ region
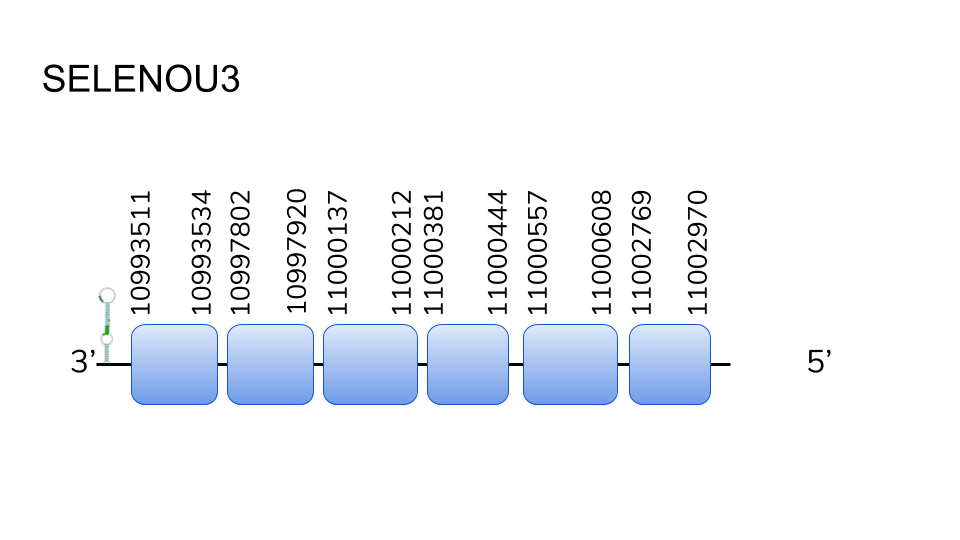
SELENOU3: Seblastian was not able to find a match for this protein, but our analysis showed 5 hits in scaffold UELW01000012.1, and the prediction process resulted in a six exon protein going from position 10993511 to 11002970, with a length of 949 base pairs. We were not able to find either a methionine starting amino acid or any selenocysteine residue, so we conclude that this might be a cystine homologue of a selenoprotein. One SECIS element was predicted in the 30 region.
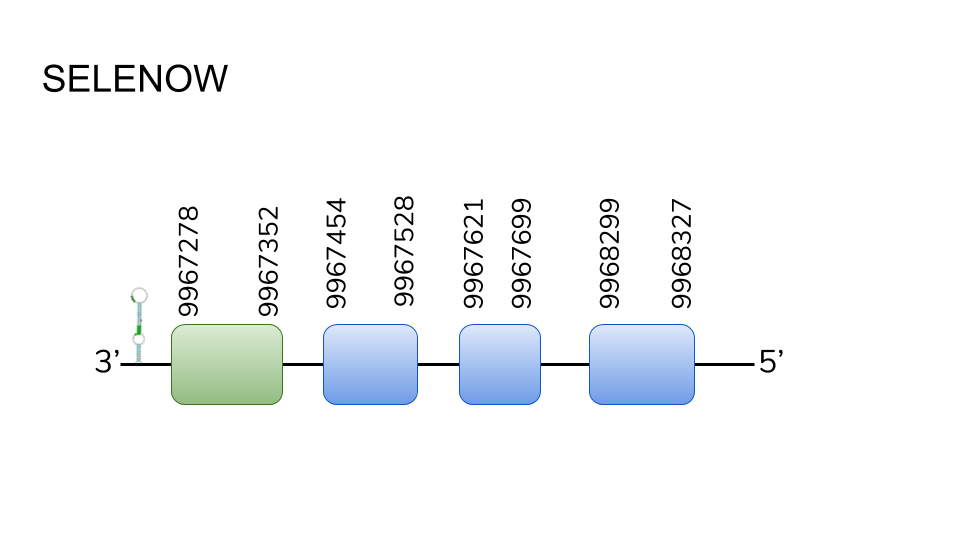
SELENOW: We found 3 hits in scaffold UELW01000522.1, and analysis though t-coffee showed high similarity and identity likeness. protein prediction resulted in a four exon protein from position 9967278 to 9968327, at a total length of 1049 exons. A selenocysteine residue was found in the first exon, but no methionine was seen at a starting point. One SECIs element was predicted in the 3’ region. The fact that we found high likeness hits, and both slenocysteine residues and SECIs elements were present makes us conclude that this protein is indeed found in Danionella dracula.
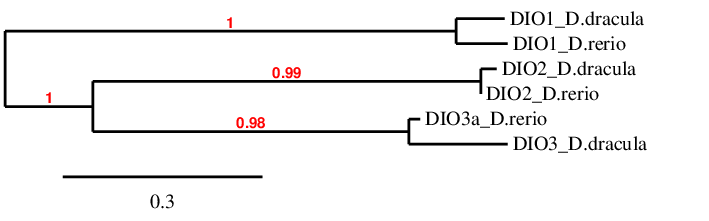
The main objective of this project was to determine and describe all selenoprotein-related genes (both selenoproteins and selenoprotein machinery) in the genome of Danionella dracula. The Zebrafish (Danio rerio) genome was used as a reference for the description of these genes. Danio rerio is a widely used species in research, and thus represents the best characterized and most studied fish genome.

