INTRODUCTION
Selenoproteins
Selenium is an essential micronutrient that is required for various redox processes and that constitutes the 21st naurally occuring amino acid: selenocystein. [1]
Selenoproteins are selenocysteine-containing proteins that have been identified throughout all 3 kingdoms of life (bacteria, archaea and eukaryota). Comparison of various animal species shows that some organisms lost the selenocysteine insertion machinery during evolution. Therefore, some selenoproteins were lost, whilst others emerged during evolutionary history.
Selenocysteine is an aminoacid encoded by UGA. This codon is currently used to terminate protein synthesis. Nonetheless, in eukaryots, a selenocysteine is inserted when a stem-loop structure (known as the selenocysteine insertion sequence or SECIS element) is located in the 3’ untranslated regions of selenoprotein genes.
Whereas a variety of organisms have been analyzed for selenoprotein occurrence, a comprehensive survey of the vertebrate or the mammalian selenoproteomes has still not been done. [2]
Synthesis of selenoproteins
Selenoproteins’s distinctive feature is the fact that they contain what is known as the 21st aminoacid: the selenocystein (Sec). This residue is encoded by the UGA codon (usually a STOP codon) that is present in the open reading frame of the gene, and it is decoded as Sec during the synthesis of ribosomal proteins thanks to specific mechanisms based on the classic translational apparatus. The main signal that will direct the recodification of the UGA codon is a secondary structure that is presented exclusively in the transcripts of selenoproteins, known as the SECIS element.
Sec is unique among other amino acids in that it is the only known amino acid in eukaryotes which biosynthesis occurs on its own tRNA, which is called tRNA[Ser]Sec. This tRNA is aminoacylated with a serine in a reaction catalyzed by the enzime seryl-tRNA synthetase (SerS) to generate the base structure for the Sec biosynthesis. Next, there is a phosphorylation of the serine by the enzyme phosphoseryl-tRNA kinase (PSTK). Selenium is in turn phosphorylated by selenophosphate synthetase 2 (SPS2), and then is added to the already phosphorylated serine. The enzyme Sec syntethase (SecS) then produces the Sec residue on this structure, forming a new structure called Sec-tRNA[Ser]Sec.[3]
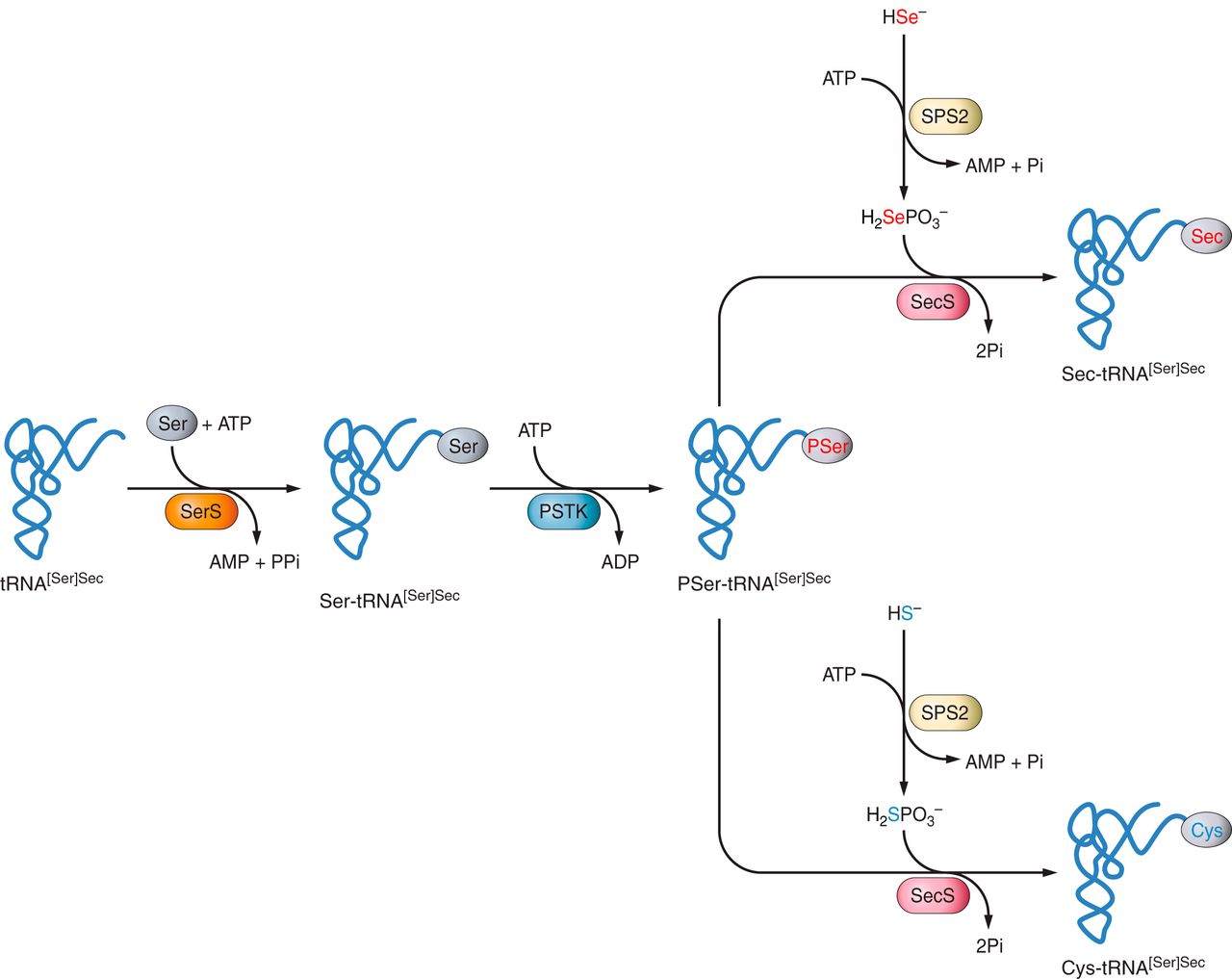
Fig 1: Mechanism of Sec biosynthesis in eukaryotes. It is shown how phosphoseryl-tRNA kinase (PSTK) provides
the phosphorylated intermediate PSer-tRNA[Ser]Sec serving as a substrate for SecS. (Lavunskyy VM et al., 2014).
In order to incorporate the Sec-tRNA[Ser]Sec into the ribosome, it must be recognized by a specific elongation factor called eEFsec. This elongation factor does not directly interact with the SECIS element, but it does so through the SECIS binding protein known as SBP2. In addition, other factors such as ribosomal protein L30 (also known as eIF4a3) participate in the process of inclusion of the Sec in response to the presence of the UGA codon in the gene reading frame.[3]
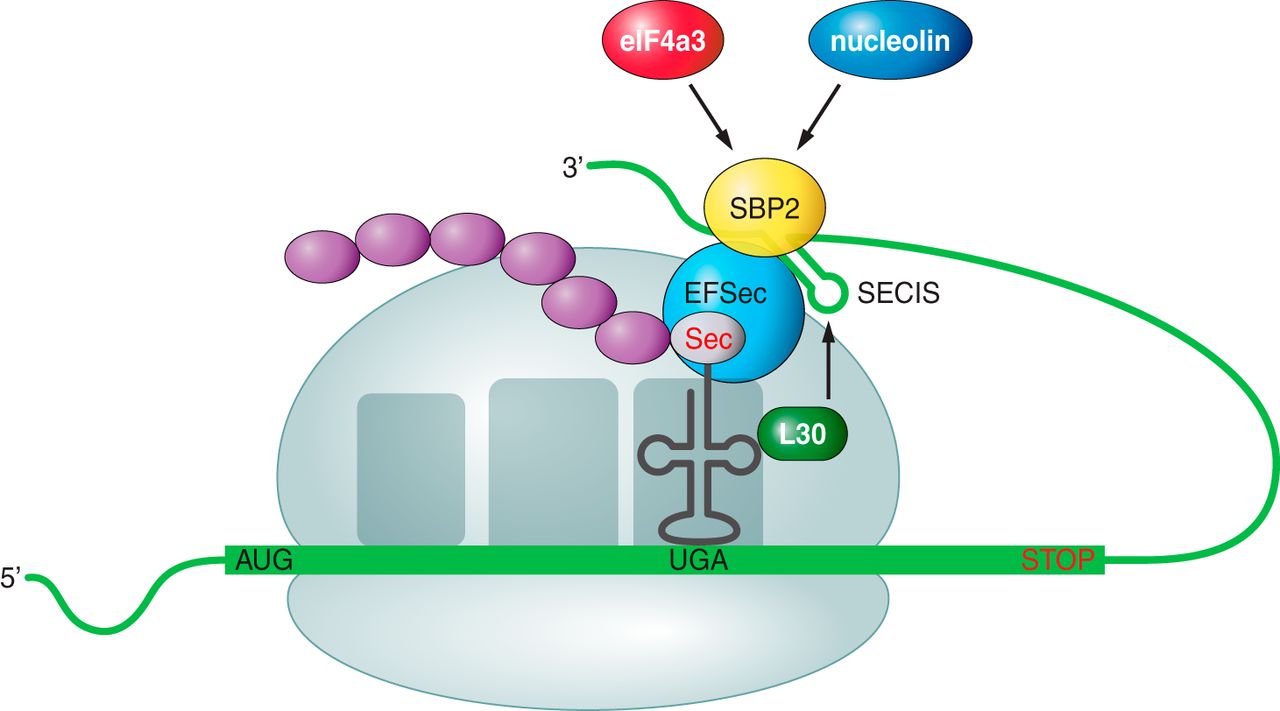
Fig 2: Mechanism of Sec insertion in eukaryotes. The figure shows known factors that are required
for Sec incorporation into proteins in response to the UGA codon (Lavunskyy VM et al., 2014).