CONCLUSIONS
In this project, we have performed an exhaustive search on the genome of Parus major in order to find the selenoproteins, the cysteine containing homologous and the essential machinery proteins for their synthesis.
For that purpose, we have automatized the process of gene prediction taking Gallus gallus proteins as a reference. Gallus gallus is not the species in SelenoDB with a closest phylogenetic relationship to Parus major as it would be Taeniopygia guttata, but as Gallus gallus is a model organism we considered that its genome would be better annotated. In addition, we have predicted the SECIs elements in the 3'-UTR region of the predicted gene of the selenoproteins and their homologous, using the SecisSearch3 software.
We expected to find the 25 selenoprotein genes that had previously been identifyed in birds (whhich were found in Gallus gallus too) in previous studies[3] and that are indicated in the following table:
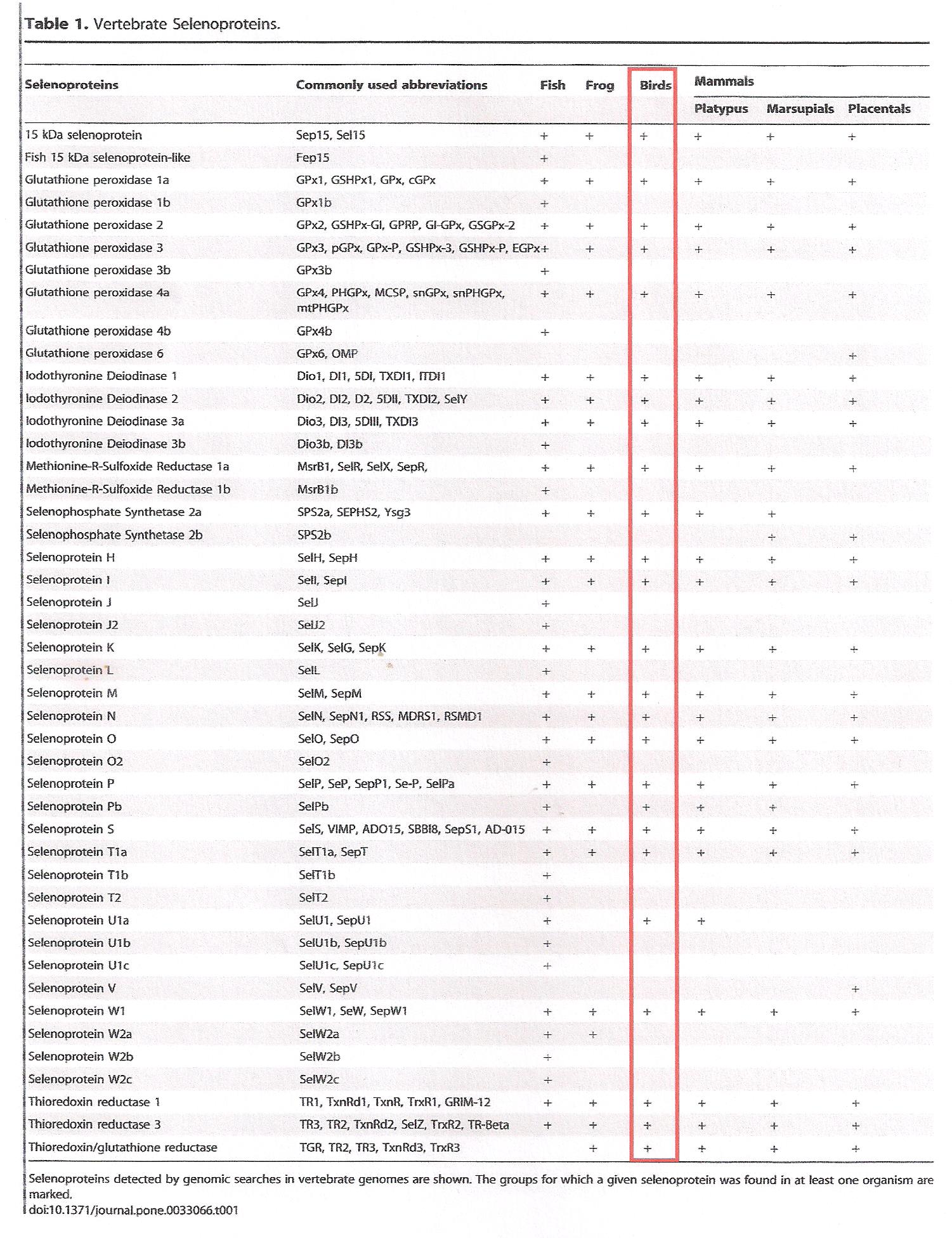
Our results suggest that Parus major does not contain all of these proteins, but contains the following ones:
- Selenoproteins: DIO1, DIO2, DIO3, GPx, GPx3, Sel15, MSRB1, SELENOK, SELENON, SELENOO1,SELENOO3, SELENOP, SELENOT, SELENOU1, TSNRD.
- Cysteine containing homologous: GPx7, GPx8.
- Machinery: eEFsec, MSRA, MSRB3, PSTK, SBP2_2, SBP2_3, SecS.
We have not found the following proteins: SBP2_1, SECp43, SELENOI, SELENOH, SELENOK_2, SELENOK_3, SBP2_1, SECp43.
Finally, we would like to point out the main limitations of our results: the genome of Parus major is fragmented in multiple contigs, a fact that difficulties the selenoprotein prediction and we cannot assure the introns and exons of the gens are properly organized. Moreover, our reference queries are proteins of Gallus gallus and this annotation may contain unknown errors. In addition, our whole project is based on the homology between Gallus gallus and Parus major genomes, without taking into account that Parus major may contain other selenoproteins, which might not be contained in the genome of Gallus gallus. Therefore, further studies should be carried out in order to be able to contrast them with our results and to better charcterize the selenoproteome of Parus major.