Introduction
Selenoproteins
A selenoprotein is a type of protein sequence peptide which incorporates one or more selenocysteine (Sec) residues. This selenocysteine, known as the 21th amino acid, has a structure which is very similar to the cysteine structure, but with an atom of Selenium instead of sulfur.[1,5,6]
 Fig 1.Selenocystein
Fig 1.Selenocystein
Selenium is very important for living organisms, because it is involved in several metabolic pathways, such as thyroid hormone metabolism, antioxidant defence system and immune function. Therefore, its absence causes diseases, but its excess is also toxic because the organism capacity of selenium chelation is overweight and free Selenium causes toxicity.[1,5,6]
Selenoproteins are found in all taxonomic levels, but there have been found some species which do not have selenoproteins in their genome.[6]
Synthesis of Selenoproteins
The synthesis of selenoproteins follows a similar process to other proteins, but because there is an incorporation of a different amino acid, selenocystein, there are several differences in the process which shall be considered.[1]
Selenium, unlike other metals, is incorporated in the polypeptide by being part of the amino acid selenocystein (Sec). Selenocystein does not have a specific codon, it is encoded by the UGA codon, which usually acts as a termination codon, but the intervention of evolutionary conserved cis- and trans- elements and protein factors allows the incorporation of a Sec to the polypeptide chain.[4,5,13]
The first step in selenoproteins synthesis is the obtention of Selenium in its selenophosphate form, which can be obtained from the diet by the action of Selenophosphate Synthetase (SPS2), a selenoprotein present in most organisms which is responsible of the conversion of selenide to monoselenophosphate, which is the active Selenium donor.[4,5,13]
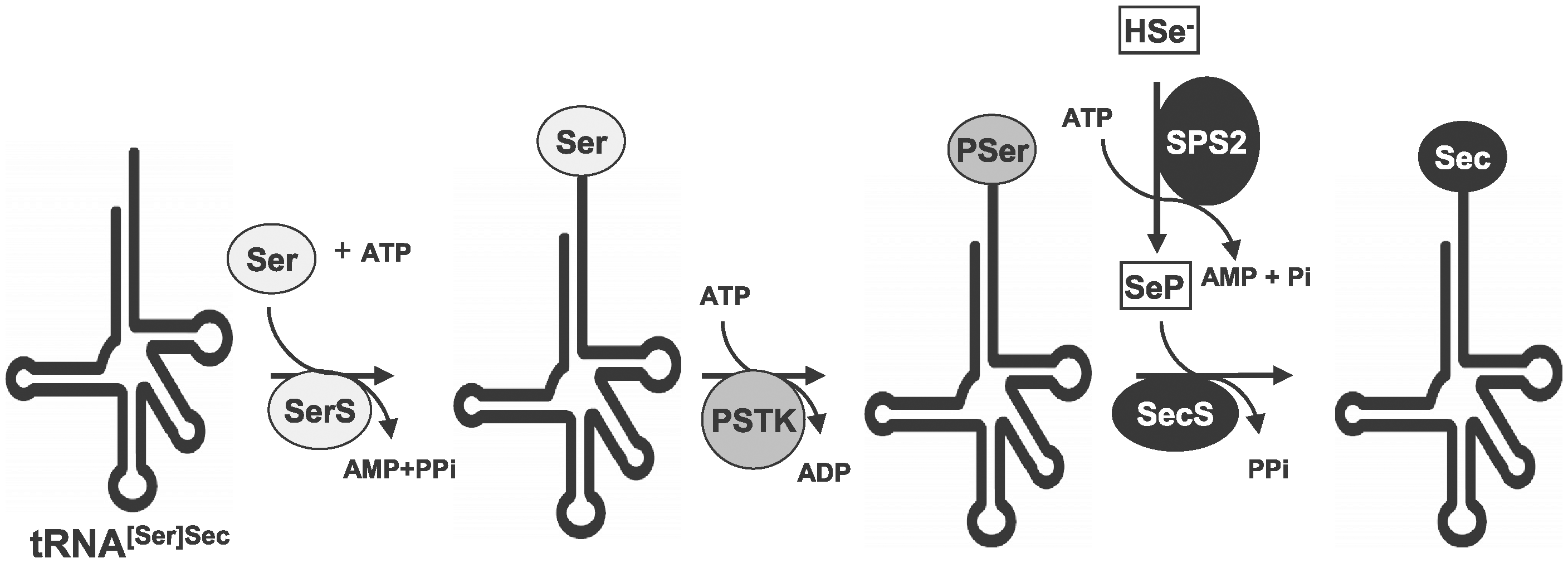
In the second place, the tRNA, responsible of the introduction of selenocystein into the polipeptidic sequence, needs to be synthesized from the serine tRNA.[4,5] A serin is attached to a Sec tRNA by a seryl-tRNA synthetase which is then phosphorylated by phosphoseryl-tRNA kinasa (PSTK). Phosphate is then replaced by the Selenium donor selenide (H2e-P) resulting into selenocysteyl-tRNA, which delivers the Sec.[4,5]
Fig 2.Selenocystein synthesis
Sec tRNA is necessary but not sufficient for decoding UGA codons as Sec. A secondary RNA stem-loop structure known as SECIS is needed. In eukaryotes, the SECIS element is usually located within the 3’-untranslated region (UTR) of the mRNAs. Its secondary structure containing consensus sequences is highly conserved showing its importance for their function.[4,5,13]
SECIS element structure includes an apical loop which contains a quartet of non–Watson-Crick G to A base-pairing forming the SECIS core preceded by an A or G residue. SECIS elements can be classified in two forms based on the existence of an additional internal loop[4,5].
However, selenoprotein synthesis machinery also includes the SECIS-binding protein 2 (SBP2), the Sec-specific elongation factor (mSelB/eEFSec) and recent findings like the ribosomal protein L30, th89e 43-kDa RNA-binding protein, Secp43, and the soluble liver antigen protein, SLA.[1,4,5]
Evolution of the Selenoproteins among species
Previous analysis of the selenoproteome in various model organisms have revealed widely different selenoproteome between organisms. For example, some algae have several selenoproteins, but higher plants and yeast do not have any. Recent studies also showed that aquatic organisms generally have a larger selenoproteome than terrestrial organisms.[6] It is also known that mammalian selenoproteomes show a trend toward reduced use of selenoproteins.
Across all vertebrates, a set of 45 selenoproteins has been identified, with at most 38 represented in a single organism (zebrafish). 27 selenoproteins were found to be unique to vertebrates.[6] 20 of them were generated through duplication of an existing selenoprotein in some vertebrate lineage, while 6 of them were part of the predicted ancestral selenoproteome. For this reason, those latter 6 proteins (GPx2, Dio2, Dio3, SelI, SelPb, Fep15) were generated at the root of vertebrates.[6] Individual mammalian selenoproteomes consist of 24/25 selenoproteins, from a set of 28. It has been shown that the mammalian selenoproteome has remained relatively stable. However, a number of evolutionary events that changed its composition were observed (Fig 3).[6]
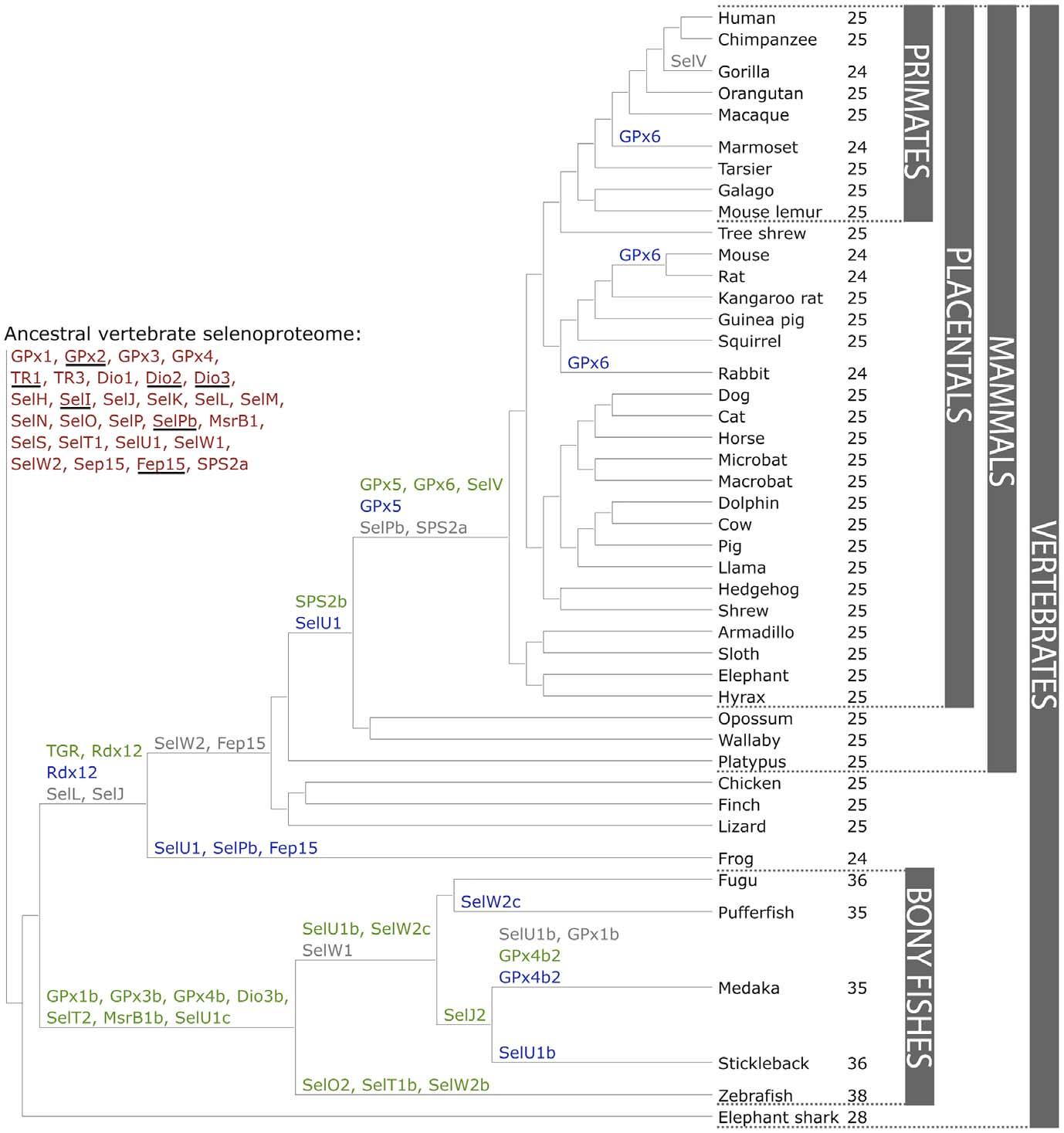
Fig 3. GPx6 and SelV were originated, SelPb was lost, SPS2b appeared and replaced SPS2a, SelV was lost in gorilla, and selenoproteins SelU1, Dio3 and GPx6 were converted to their cys-containing forms in major or minor mammalian lineages.
Neotoma lepida
Neotoma lepida, commonly known as desert woodrat, is a species of pack rat native of desert regions of western North America. These animals are relatively small, as their length varies between 27 and 40cm including a considerably long tail.[14] They can present a wide range of pelage colouring on the back, but all the individuals present white underside and feet. The tails of Neotoma lepida individuals are always bicolored.[14] There are not important differences between male and female.[14]
They predominantly eat leaves and grain, being able to eat other types of food when necessary.[14]
Neotoma lepida breading season is between October and May. Gestation periode takes between 30 and 36 days.[14] Weening occurs between 27 and 40 days after birth, and the cubs reach sexual maturity after 2 to 3 months.[14]
Taxonomic classification:
- - Domain: Eukarya
- - Kingdom: Animalia
- - Philum: Chordata
- - Class: Mammalia
- - Order: Rodentia
- - Superfamily: Muroidea
- - Family: Cricetidae
- - Subfamily:Neotominae
- - Genus: Neotoma
- - Species: N. lepida
Reference genome
Mus musculus
We have chosen Mus musculus genome and so, their selenoproteins, because it belongs to the same order, Rodentia, as Neotoma lepida, and because of that we can suspect that there will be a strong conservation of the selenoproteins between the two genomes. Mus musculus is one of the major organisms for comparative genome analysis, and this is important in order to get proper alignments.[9]
Sequentiation of 2.6Gb of this genome have already been described with less than one sequencing error per 10-5 base pairs. A few chromosomes (Chr 2, 4, 11 and X) are entirely sequenced, allowing comparisons with homologous regions of other genomes.[3,9]
Regarding selenoproteins, Mus musculus has a total of 24 along its genome.[6]
Homo sapiens
We have chosen Homo sapiens genome and so, their selenoproteins, because is the best characterized genome and also is a mammal, so it is a really good alternative for Rodentia species.[8]
Such genome, has a length of 3 Gb and is conformed by 23 pairs of chromosomes.[8] Regarding the selenoproteins, Homo sapiens has a total of 25 selenoproteins along its genome.[6,8]
Rattus norvegicus
We have chosen Rattus norvegicus genome and so, their selenoproteins, because it belongs to the same order, Rodentia, as Neotoma lepida, and because we suspect, like in Mus musculus, that there will be a strong conservation of the selenoproteins between the two genomes. Although it is not such a good model as Mus musculus, its genome is well characterized, so it is a viable alternative for those proteins which could not be characterized when comparing our genome with Mus musculus.[10]
Rattus norvegicus genome has a length of 48,7Mb and is conformed by 21 pairs of chromosomes.[10] Regarding the selenoproteins, Rattus norvegicus has a total of 24 selenoproteins along its genome.[6,10]
|