Selenoproteins are a group of proteins which contain a residue of selenocysteine (Sec), known as the 21st amino acid. This unusual amino acid is a cysteine analog with selenium instead of sulfur in the radical chain. Interestingly, Sec is cotranslationally inserted into the emergent polypeptide in response to UGA codons (Mariotti et al, 2010) which usually acts as a STOP codon. This dual function of UGA codon has been ignored for many years, with the subsequent misinterpretation of UGA codon as Sec codon and terminator.
The inclusion of the Sec and the synthesis of a selenoprotein require a sequence in the 3'-UTR region called SElenoCysteine Insertion Sequence (SECIS). The eukaryotic SECIS element has a length of approximately 60 nucleotides and it adopts a stem-loop structure in the mRNA.
From an evolutionary point of view, the maintenance of a selenoprotein is a trade-off between its efficacy and the handicap of synthethising it. Some selenocysteines mutate to cysteine but keep their functionality. Thus, some members of selenoprotein families are analogs containing cysteine.
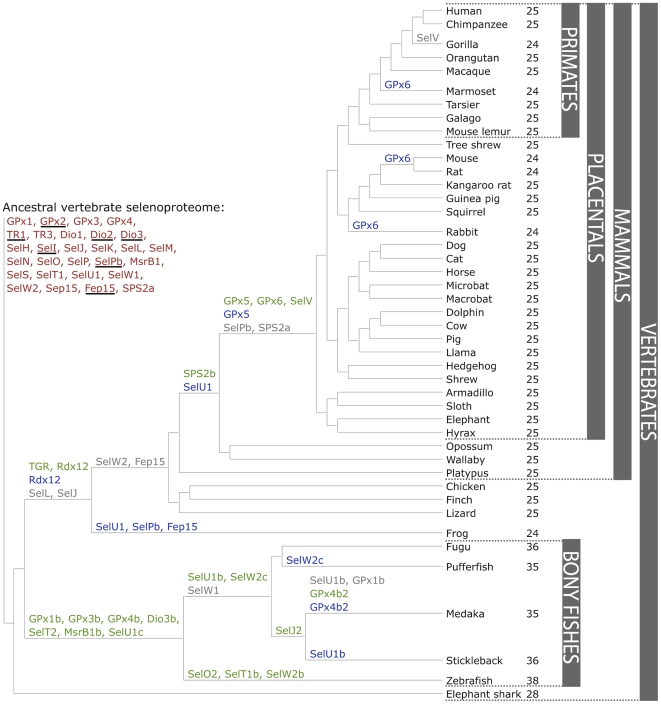
The first comprehensive analysis of all the selenoprotein families in mammals was published very recently (Mariotti et al., 2012). This survey of several mammalian genomes gives us a wide view of the families present in mammals and their changes along evolution and separation of clades.
There is great number and a vast variety of selenoproteins. To date, 45 selenoprotein families have been described (Mariotti et al, 2012). Bony fishes have the largest variety with 41 different families, whereas mammals have 28 families all together. According to previous studies a single mammalian species can have up to 25 selenoproteins in its genome.
Perhaps due to their complexity, a lot about selenoproteins is still unknown. Several families do not have an assigned function, and the distribution of selenoproteins among the evolutionary tree is incomplete. Moreover, new proteins involved in UGA recoding have been identified, although their function is not yet known.
Nowadays a lot of new organisms are being genotyped, but the distribution of selenoporoteins in their genomes is unknown. Expanding the number of genomes with annotated selenoproteins would provide a better understanding of their evolution. From this we could derive their importance, and be closer to inferring their function and their implication in disease.
Synthesis of selenoproteins
The common feature of selenoproteins is the fact that they contain at least one residue of selenocysteine. Selenocysteine is an aminoacid which is analogous to cyestine but contains an atom of selenium. This aminoacid is coded by the codon UGA, which would usually code for a STOP.
The UGA codon duality is circumvented by the presence of conserved cis- and trans- acting elements dedicated to the decoding of UGA as a SeCys residue. (Allmang et al, 2006; Lobanov et al, 2009; Castellano et al, 2009).
The basic cis-acting elements for the synthesis of a selenoprotein is the SECIS element, as well as the in-frame UGA codon. On the other hand, the basic trans-acting elements are SPS1,SPS2, SecS, Pstk, eEFsec, SBP2 and SeCys/tRNA[Ser]Sec. All these factors are globally known as the selenoprotein translation machinery.
Interestingly, Sec is the only aminoacid synthesized directly on the tRNA, and isn’t produced from a cysteine but from a serine. tRNA Pstk phosphorylates the tRNA[Ser]Sec , which will allow the next reaction. Selenophosphate synthetase 2 SPS2 prepares the Selenium to be incorporated, and selenocysteine synthase (SecS) binds the atom to the serine, completing the synthesis of the tRNA. This tRNA is not recognized by usual elongation factors, and is instead bound to the specific factor eEFSec.
tRNA Selenocysteine 1 associated protein 1 (Secp43) is also involved in the synthesis of the tRNA[Ser]Sec and selenoproteins (Reeves et al, 2009).
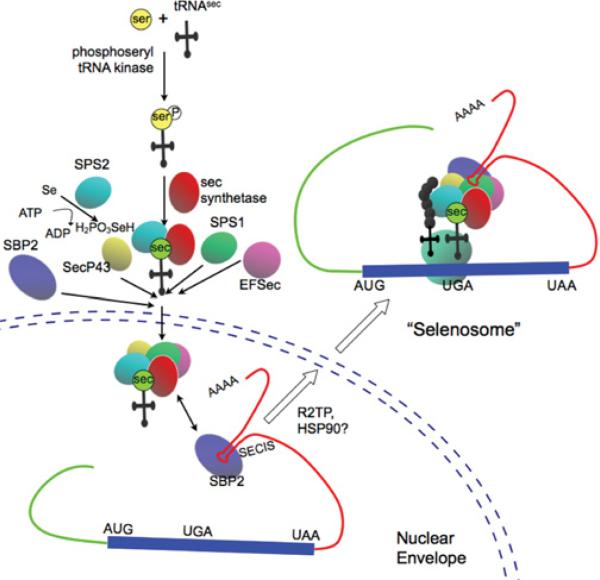
Image: Machinery involved in synthesis of selenoproteins (Bellinger F.P., et al; 2009).
After the trans-acting elements have participated in the tRNA synthesis, the SECIS element acts in Cys to recode the UGA codon. SECIS recruits SECIS Biding Protein 2 (SBP2). SBP2 will in turn recruit and bind eEFSec, the specific elongation factor. Finally to achieve the incorporation of the selenocysteine, the protein has to select the tRNA[Ser]Sec, which is taken to the UGA codon.
We can distinguish two major groups of selenoproteins depending on their function. The first group is formed by housekeeping genes and the second group contains stress activated proteins.