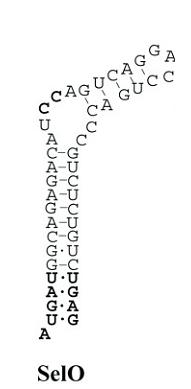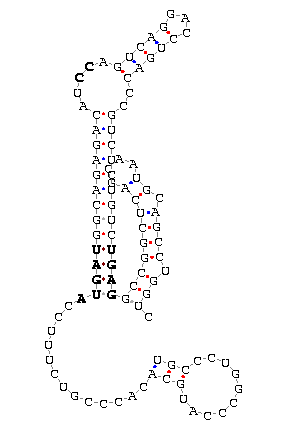RESULTS & DISCUSSION
We will explain the results for each selenoprotein on which we have been working.
Selenoprotein I
Mapping:
We obtain the transcript report of this selenoprotein from the ensembl database.
The genomic location is on chromosome 2p23.3, between the 26480633 and 26530402bp.
The gene contains 10 exons that are transcribed into 8093bp on mRNA and the protein result is composed by 397 residues. The selenocysteine is located in the residue 387. SELi mRNA was detected in a variety of tissues and cell types.
All the information about the mapping is resumed in this gff.
Finding SNPs:
We can extract from the transcript report all the SNPs located in the sequence, where they are and which kind of nucleotide change makes a polymorphism.
We have found so many SNPs but we are only interested in those which affect the SecIS element, the SNPs that are in the selenoprotein or close to it.
We obtained a complete list of all the SNPs that mostly are contained in introns.
There are two SNPs affecting the protein: a T or G in the residue 102, and a T or C in the residue 289, but both are synonymous changes so the aminoacid maintains conserved. As the change produced is synonymous there is no alteration in the peptide sequence of the selenoprotein and therefore we will not study it.
| Residue | SNP ID | SNP type | Alleles | Ambiguity code | Alternate residues |
| 102 | rs6748996 | Synonymous | T/G | K | - |
| 299 | rs934280 | Synonymous | T/C | Y | - |
Table 1: SNPs from the the selenoprotein I.
The SNPs located in the 3’UTR region of the mRNA do not affect the SecIS structure, so we will not study them.
Analyzing SecIS elements structure:

With the 3’UTR sequence obtained thanks to the possibility of extracting data in fasta files, we can use the SECISarch program.
We past the sequence and we obtain the SecIS structure predicted in our sequence. The structure of the SecIS is correct and it has not any strange form or nucleotide pattern.
The program also give us the SecIS sequence, which will be used in the next studies.
Figure 9: SecIS structure from Sel I.
Analyzing SecIS elements:
In this step we made use of blastn. As we have explained previously we blast our SecIS sequence with the est_human database.
The results show the alignment of our SecIS sequence with different database est sequences. The blastn output shows the identifier, the identity percent between the query sequence (our sequence pasted) and subject sequence (est sequences from the database), and the align of both sequences.
This output of the blastn was run in the fndsnp.pl that we have done, and we obtain the following output.
The program selects the aligns that contain an identity percent between 95 and 99%. So we have now a selection of the subject sequences that came from the est_human database.
We can see there is a sequence that contains only one SNP from the output file. Moreover the program produces a list of all SNPs that have been found in all sequences, and it counts how many times each change has occurred.
So we can see which the most habitual SNPs are. In this case, we have only found one SNP at the
position 18 consisting in a T or a gap, counting from the begining of the SecIS sequence.
Analyzing SecIS elements with SNPs:
Now we know the SNP, but we want to analyze how this SNP affects the SecIS structure.
So we prepared the Obtain_SecIS_sequence program to obtain our query sequence (SecIS sequence of the Sel I) with the SNP on it.
We will find this sequence in the output file.
This output is introduced into the SECISearch program to be able to obtain the SecIS Structures.
The result shows us that the Secis has a good structure and that the SNP does not affect the basic structure or the base-pairing patterns. Only a mutation in this motif: ATGA_GA could affect in an important way the SecIS structure.
Relation between SecIS sequence and diseases:
The following table sumarises all the information mentioned above.
| ID Est | Origin | SNPs | SecIS Alteration |
| gi|11984132|gb|BF698724.1| | 602126262F1 NIH_MGC_56 Homo sapiens cDNA clone | 18,T/- | NO |
Table 2: SNP from the SecIS element of selenoprotein I.
No relation has been found between the Selenoprotein I and some kind
of disease in current databases.
As we have seen above, the position where the SNP is located is in the nucleotide 18 counting from the begining
of the SecIS sequence. The SNP produces a T or a gap in this position, as the table shows. This est sequence comes from a
primitive neuroectoderm, and the SecIS element is not altered.
These facts lead us think there is no relationship between selenoprotein I and any
disease.
Selenoproteina N
Mapping:
We have found in the ensembl database two transcripts of this protein.
So we have one trasncrpit report for the first transcript and another one for the second trascript.
The genomic location is on chromosome 1p35-36, between the 25810868 and 25828856bp.
The two transcripts that we have found are two different isoforms and differ in the fact that the transcript 1 contains one more exon (number 3) than the transcript 2, where it has been spliced out.
Moreover a selenocysteine is located in this exon.
So the transcript one contains one more exon and has 2 selenocysteines while the transcript 2 contains one exon less and only one selenocysteine.
Ensembl database contains some errors related to the isoform 2. We find out that the isoform 2 has one selenocystein at the same position as the isoform 1.
In the residue 428 and 429 of the isoform 2, we find a Histidine and a Cysteine instead of a Selenocysteine and a Glycine.
This finding is supported by some pubmed articles (1,2,3), where we have found that isoform 1 has two selenocysteines.
Moreover the first one (located in the exon 3, spliced out in the isoform 2) can code for a codon stop resulting a shorter peptid.
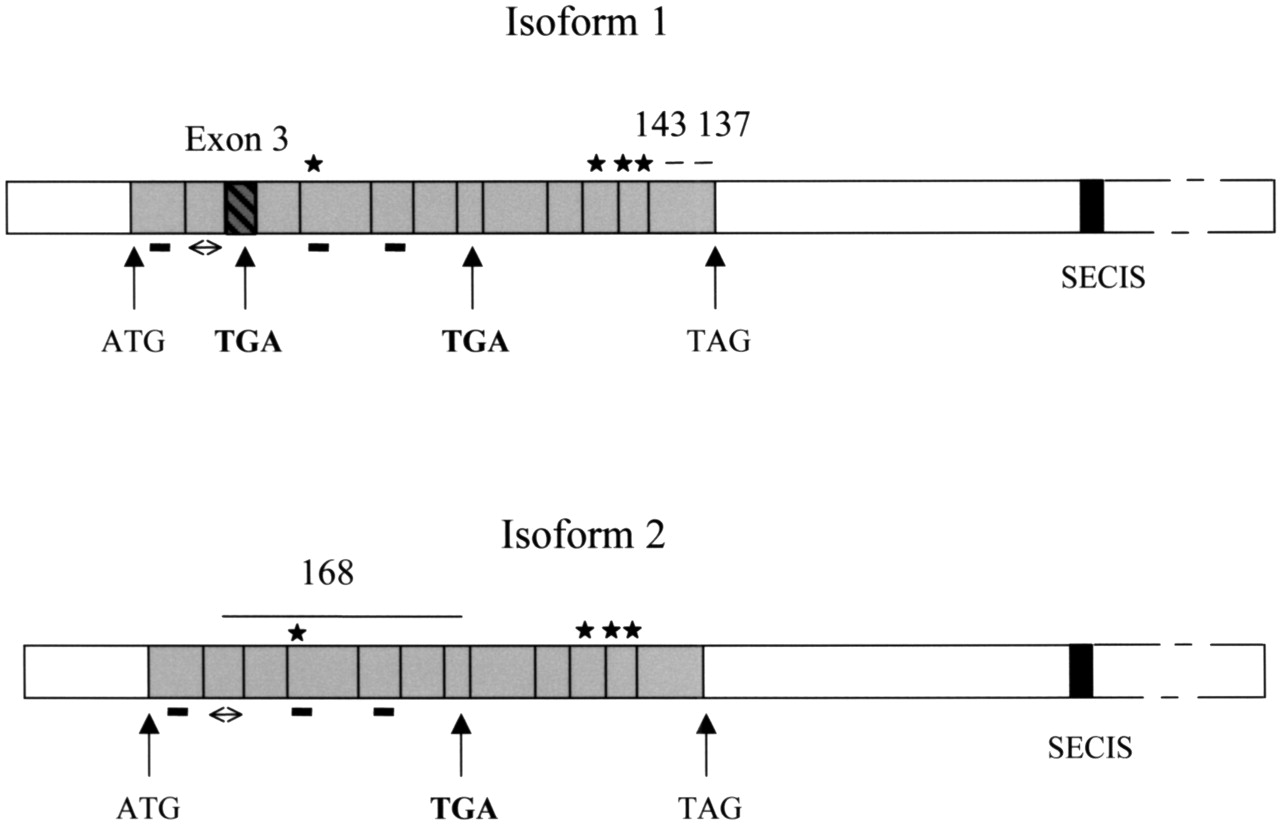
Figure 10: Isoforms of Sel N
The gene contains 13 exons that are transcribed into 4274bp on mRNA and the protein result is composed by 590 residues in the isoform 1 and by 556 residues in the isoform 2.
The selenocysteine are located in the residue 127 and 462 in the isoform 1 and in the residue 428 in the isoform 2.
SEPN is present at a high level in several human fetal tissues and at a lower level in adult tissues, including skeletal muscle. Both transcripts were detected in skeletal muscle, brain, lung, and placenta,
but isoform 2 was always predominant.
All the information about the mapping of the transcripts is resumed in this gff .
Finding SNPs:
We can extract from the transcript report all the SNPs located in the sequence, where they are and which kind of nucleotide change makes a polymorphism.
We have found so many SNPs but we are only interested in those which affect the SecIS element, the SNPs that are in the selenoprotein or close to it.
We obtained a complete list of all the SNPs from isoform 1 and isoform 2.
| Residue | SNP ID | SNP type | Alleles | Ambiguity code | Alternate residues |
| 106 | 1132855 | Non-synonymous | A/G | R | A, T |
| 142 | 7349185 | Non-synonymous | G/A | R | C, Y |
| 391 | 760597 | Synonymous | T/C | Y | - |
| 443 | 12023720 | Synonymous | G/A | R | - |
| 502 | 2294228 | Non-synonymous | C/A | M | N, K |
Table 3: SNP from the SecIS element of the isoform 1 selenoprotein N.
| Residue | SNP ID | SNP type | Alleles | Ambiguity code | Alternate residues |
| 108 | 7349185 | Non-synonymous | G/A | R | C, Y |
| 357 | 760597 | Synonymous | T/C | Y | - |
| 409 | 12023720 | Synonymous | G/A | R | - |
| 468 | 2294228 | Non-synonymous | C/A | M | N, K |
Table 4: SNP from the SecIS element of the isoform 2 selenoprotein N.
None of the SNPs found are in the SecIS structure, close the selenocysteine,
or affecting this aminoacid, therefore we are not interested on them.
Based on fact that two isoforms come from the same gene but by alternative splicings, both isoforms
should contain the same aminoacid sequence but one of them containing one more exon.
As we have said before there is a difference between two isoforms in the aminoacid sequence that
they share.
Surprisingly, ensembl does not find any SNP in the codons of the two aminoacids that differ from one
isoform to the other one. This means that ensembl database contains errors or unespicified facts.
Analyzing SecIS elements structure:
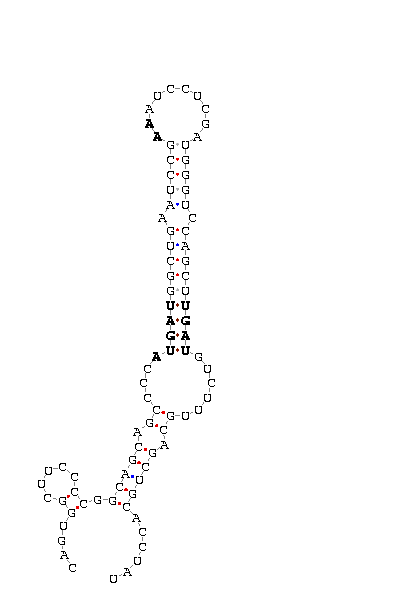
With the 3’UTR sequence obtained thanks to the possibility of extracting data in fasta files, we can use the SECISearch program.
We past the sequence and we obtain the SecIS structure predicted in our sequence. The program shows the different SecIS structure elements, they are correct and the base-pairing patterns seem to be also correct.
Figure 11: SecIS structure from Sel N.
Analyzing SecIS elements:
The output of the blastn was run in the findsnp.pl program that we have prepared, and we obtain the output.
As we have made with the selenoprotein I, we obtain a list with the different sequences with the SNPs and also a number of the different SNPs met.
We found 10 sequences with SNPs from 99 Blast hits on the Query Sequence; we counted 16 different SNPs, but only appearing once.
Analyzing SecIS elements with SNPs:
To evaluate the different SNPs that we have met, we use Obtain_SecIS_sequence program to obtain our query sequence (SecIS sequence of the Sel N) with one polymorphism in it. With this program we obtain a list of all the sequences each one with a SNP in an output.
With this output we go to the SECISearch and we obtain the different SecIS structures each one for each sequence introduced.
The SNPs seems not affect the structure of the SecIS element, all seems to be correct and no alteration will appear.
| SNP | Frequence | Affects Structure? | SNP | Frequence | Affects Structure? |
| 12,C/G | 1 | NO | 52,T/A | 1 | NO |
| 14,G/T | 1 | NO | 57,C/A | 1 | NO |
| 14,-/C | 1 | NO | 61,-/C | 1 | YES?? |
| 16,-/T | 1 | NO | 70,-/T/C/A/G | 1 | NO |
| 17,-/G | 1 | NO | 72,T/C | 1 | NO |
| 21,G/A | 1 | NO | 73,-/G | 1 | NO |
| 25,C/- | 1 | NO | 75,-/A | 1 | NO |
| 43,A/G | 1 | NO | 82,A/C | 1 | NO |
Table 5: Appearance frequency of each SNP.
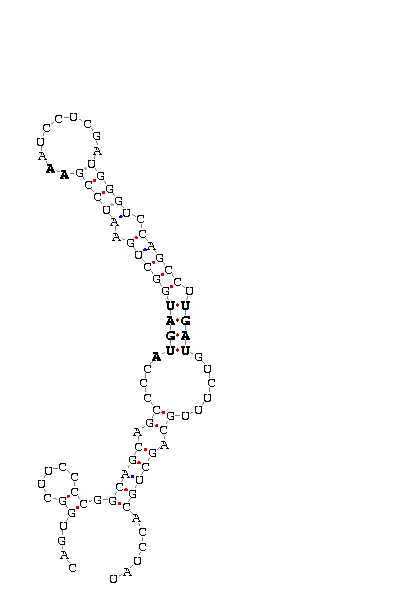
Only the SecIS from the est with the id: gi|18797332|gb|bm556239.1|,
can present some kind of problems because of the SNP located in the residue 61 from the begining
of the secis sequence (61-c: 1 87), next to the important motif described above. This nucleotide
does not match with anything, therefore this fact can produce an alteration of secis function and
the selenoprotein in general.
Figure 12: Secis structure from Sel N adding a C at position 61.
With the Obtain_SecIS_sequence program2 that we have prepared,
we can obtain each subject sequence with all the SNPs that it contains. We have sequences with only one SNP but we have others with more than one,
and more than one possibility of nucleotide change due to the iupac ambiguity code where each letter can correspond to more than one nucleotide.
So with this program we obtain the sequences chosen from the blast output with all the SNPs that contain.
Now with this output file of our third program we use again the SECISearch program to analyze the SecIS structures of these sequences.
| ID Est | Origin | SNPs | Secis Alteration |
| gi|15164992|emb|AL601486.1| | DKFZp313N1642_r1 313 (synonym: hlcc2) Homo sapiens cDNA clone DKFZp313N1642 5' | 70,-/A/C/G/T
| NO |
| gi|18797332|gb|BM556239.1| | AGENCOURT_6544460 NIH_MGC_88 Homo sapiens cDNA clone IMAGE:5550208 5' | 61, -/C | YES? |
| gi|16771388|gb|bm042121.1| | 603615746F1 NIH_MGC_112 Homo sapiens cDNA clone IMAGE:5420674 5' | 12, C/G; 43, A/G | NO |
| gi|10160532|gb|be746540.1| | 601580105F1 NIH_MGC_9 Homo sapiens cDNA clone IMAGE:3928800 5' | 57, C/A; 73, -/G | NO |
| gi|14294011|gb|bg913535.1| | NCI_CGAP_Brn67 Homo sapiens cDNA clone IMAGE:4943421 5' | 14,G/T;16,-/T;17,-/G | NO |
| gi|845754|gb|r71722.1| | yj85g05.r1 Soares breast 2NbHBst Homo sapiens cDNA clone IMAGE:155576 5' | 25,C/- | NO |
| gi|16523466|gb|bm009112.1| | 603629475F1 NIH_MGC_41 Homo sapiens cDNA clone IMAGE:5434747 5' | 75,-/A | NO |
| gi|22694159|gb|bu180175.1| | GENCOURT_8107027 NIH_MGC_112 Homo sapiens cDNA clone IMAGE:6267459 5' | 21,G/A; 52,T/A | NO |
| gi|19362988|gb|bm912609.1| | GENCOURT_6612214 NIH_MGC_41 Homo sapiens cDNA clone IMAGE:5474167 5' | 14,-/C; 82A/C | NO |
| gi|10204277|gb|be783079.1| | 601470665F1 NIH_MGC_67 Homo sapiens cDNA clone IMAGE:3873688 5' | 72,T/C | NO |
Table 6: SNPs from the secis element of the selenoprotein N.
The result shows us different SecIS structures each one to each sequence. All of them seem to be normal SecIS, and none has alterations
in the important motif (ATGA_GA ) for the SecIS structure. Moreover the score of all the SecIS structures calculated by the SECISearch program are very solid.
Relation between SecIS sequence and diseases:
All the est sequences found in the blast search hits
come from RNA of no altered cells, so the sequence that we have mentioned above
that could implicate an alteration in the secis structure has no relevance.
The association between selenium deficiency and muscular dystrophy in
livestock suggests a role for selenium in the pathophysiology of striated muscles.
Selenoprotein N has been associated to muscular dystrophy, causing congenital
muscular dystrophy with spinal rigidity and restrictive respiratory syndrome by mutations
in SEPN1.
Selenoprotein N is high expressed in cultured myoblasts, and is also downregulated
in differentiating myotubes, suggesting a role for SEPN1 in early development and in cell
proliferation or regeneration.
It is one the first descriptions of the relation of a selenoprotein implicated in a human
disease.
According to OMIM, there are many different allelic variants associated to the SEPN
and to the spine muscular dystrophy. Two of the allelic variants are related to changes
in a selenocysteine.
One for a T-to-G transversion at nucleotide 1384 in exon 10 of the SEPN1 gene, resulting
in a sec462-to-gly (U462G) substitution. The second is a G-to-A transition at nucleotide 1385
of the SEPN1 gene, in this case the mutation changes the selenocysteine (sec) codon (TGA) into
a stop codon (TAA), preventing selenocysteine incorporation and leading to a shorter protein.
Selenoproteina O
Mapping:
The genomic location is on chromosome 22q13.33, between the 48941865 and 48958501bp.
The gene contains 9 exons that are transcribed into 2314bp on mRNA and the protein result is composed by 669 residues. The selenocysteine is located in the residue 667.
SELO mRNA was detected in a variety of tissues and cell type.
All the information about the mapping is resumed in the gff.
Finding SNPs:
We can extract from the transcript report all the SNPs located in the sequence, where they are and which kind of nucleotide change makes a polymorphism.
We have found so many SNPs but we are only interested in those which affect the SecIS element, the SNPs that are in the selenoprotein or close to it.
We obtained a complete list of all the SNPs.
| Residue | SNP ID | SNP type | Alleles | Ambiguity code | Alternate residues |
| 3 | 5771225 | Non-synonymous | T/C | Y | A, V |
| 21 | 5771226 | Synonymous | T/C | Y | - |
| 57 | 6010201 | Synonymous | C/G | S | - |
| 105 | 3747942 | Synonymous | T/C | Y | - |
| 167 | 2272846 | Non-synonymous | C/A | M | T,N |
| 216 | 5771231 | Non-synonymous | G/T | K | S, A |
| 630 | 2272852 | Non-synonymous | G/A | R | K,E |
| 638 | 17013238 | Non-synonymous | G/A | R | K, E |
| 640 | 9617118 | Synonymous | G/A | R | - |
| 658 | 1052827 | Synonymous | C/T | Y | - |
| 669 | 1052832 | Non-synonymous | C/T | Y | S,L |
Table 7: SNPs from the secis element of the selenoprotein O.
None of the SNPs found are in the SecIS structure, or close the selenocysteine, so they are not relevant for our project.
Analyzing SecIS elements structure:
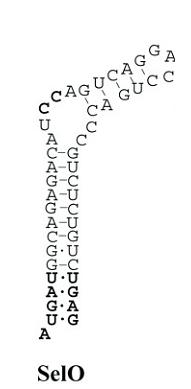
With this selenoprotein we have had some problems in obtaining the
structure of the SecIS element. SECISearch program used with the other selenoproteins can not be
used now in this case because the program does not find any secis structure. Our tutor gave us a
new program that detects the secis sequence in the 3'UTR sequence of the selenoprotein with a different pattern,
evaluates the thermodynamic energy and shows at the end the secis structure. RNAFold and patscan program are also needed to run this program.
We obtained the SecIS structure predicted in our sequence. The program shows the SecIS structure for our sequence and it seems to be correct and nothing strange neither in the structure nor in the base-pairing pattern.
Figure 13: Secis structure from Sel O.
Analyzing SecIS elements:
The output of the blastn was run in the findsnp.pl program that we have done, and we obtain the output. As we have made with the other two selenoproteins we obtain a list with the different sequences with the SNPs and also a number of the different SNPs met.
We found 14 sequences with SNPs from 59 Blast hits on the Query Sequence; we counted 22 different SNPs, some appearing once and
the other more than one time. If some SNPS appear more than once in tissues we can expect
there is a relationship between these two facts.
Analyzing SecIS elements with SNPs:
Thanks to Obtain_SecIS_sequence program,
we can prepare a list of all the sequences only with one SNP in an output.
Going to the SECISearch we obtain the different SecIS structures each one for each sequence introduced that contains one SNP.
| SNP | Frequence | Afects Structure | SNP | Frequence | Afects Structure |
| 4,C/A | 2 | NO | 72,C/T | 5 | NO |
| 5,C/T | 1 | NO | 73,-/G | 1 | NO |
| 39,-/T | 1 | YES | 87,-/T | 1 | NO |
| 39,G/A | 3 | YES | 86,G/A | 3 | NO |
| 45,T/A | 1 | NO | 89,G/A | 1 | NO |
| 53,G/C | 1 | YES | 90,T/A | 1 | NO |
| 56C/- | 1 | NO | 94,G/A | 1 | NO |
| 66,G/T | 3 | NO | 95,C/_ | 1 | NO |
| 68,C/T | 3 | NO | 98,G/C/T/A | 1 | NO |
| 70-/C | 1 | NO | 100,T/A | 1 | NO |
| 71,T/A | 1 | YES |
Table 8: Appearance frequency of each SNP.
We have not been able to obtain the different images from each SecIS structure due to
an informatic problem, but we have obtained the SecIS sequence and we are able to know if the
SecIS structure can occur or not. We have analyzed 24 different sequences, but we only have
results for 20 of them, so the SecIS element is only viable in 20 of them. These 4 sequences that
seem not to have a SecIS element can have some alterations in their sequence affecting the
matches necessaries to form the shape of the SecIS element. For instance, these alterations can
be in the relevant motif (ATGA_GA) or we can find an unpaired nucleotide.
The third program (Obtain_SecIS_sequence2 program), now applied to Sel O,
makes an output file.
This file sequences with more than one SNP.
Pasting the sequences in the SECISearch program we can analyze the SecIS structures of these sequences.
The result shows us the different secis structures obtained.
We can see there are less secis structures in the result obtained than sequences introduced into the SECISearch program due
to the fact that there should be repeated sequences and also secis structures with no
thermodinamic stability that are not shown in the results of this program.
The SecIS structures are correct and nothing has happened in the structure so they are correct and viable.
| ID Est | Origin | SNPs | Secis Alteration |
| gi|46569321|emb|bx368938.2| | BX368938 Homo sapiens HELA CELLS COT 25-NORMALIZED cDNA clone CS0DK012YM07 5' | 71,T/A;95,C/A;100,T/A | NO |
| gi|10333084|gb|be884308.1| | 601505777F1 NIH_MGC_71 Homo sapiens cDNA clone IMAGE:3907314 5' | 56,C/-; 95,C/- | NO |
| gi|6141147|gb|aw137014.1| | UI-H-BI1-acu-d-08-0-UI.s1 NCI_CGAP_Sub3 Homo sapiens cDNA clone IMAGE:2715686 3' | 45,T/A; 53,G/C; 94,G/A | NO |
| gi|46554667|emb|bx354872.2| | BX354872 Homo sapiens NEUROBLASTOMA COT 25-NORMALIZED cDNA clone CS0DC026YN23 3' | 4,C/A; 89,G/A; 90,T/A | YES?? |
| gi|3694022|gb|ai160642.1| | qc89a02.x1 Soares_pregnant_uterus_NbHPU Homo sapiens cDNA clone IMAGE:1721354 3' | 72,C/T; 86,G/A; | NO |
| gi|46304542|emb|bx354948.2| | BX354948 Homo sapiens NEUROBLASTOMA COT 25-NORMALIZED cDNA clone CS0DC027YH03 3' | 4,C/A | NO |
| gi|3213814|gb|ai004304.1| | ou56e04.x1 NCI_CGAP_Br2 Homo sapiens cDNA clone IMAGE:1631838 3' | 39,G/A;66,C/T;68,C/T;72,C/T;86,G/A | NO |
| gi|20135238|gb|bq102254.1| | ij19a02.x1 Melton Normalized Human Islet 4 N4-HIS 1 cDNAclone IMAGE:6135051 3' | 66,C/T;68,C/T;72,C/T | NO |
| gi|46307094|emb|bx349459.2| | X349459 Homo sapiens NEUROBLASTOMA COT 25-NORMALIZED cDNA clone CS0DC027YM11 3' | 70,-/C;73,-/G | YES?? |
| gi|5056776|gb|ai735252.1| | at08c08.x1 Barstead aorta HPLRB6 Homo sapiens cDNA cloneIMAGE:2354510 3' similar to SW:YDIU_ECOLI P77649 HYPOTHETICAL 54.4 KD PROTEIN IN AROH-NLPC INTERGENIC | 39,G/A;72C/T | NO |
| gi|14168296|gb|bg820709.1| | 602780639F1 NCI_CGAP_Brn67 Homo sapiens cDNA clone IMAGE:4931528 5' | 39,-/T; 87,-/T | NO |
| gi|6144075|gb|aw139357.1| | UI-H-BI1-ada-g-12-0-UI.s1 NCI_CGAP_Sub3 Homo sapiens cDNA clone IMAGE:2716246 3' | 5,C/T | NO |
| gi|5673006|gb|ai934136.1| | wn97e01.x1 NCI_CGAP_Ut1 Homo sapiens cDNA clone IMAGE:2453784 3' | 39,G/A;66,C/T;68,C/T;72,C/T;86,G/A | NO |
| gi|46304039|emb|bx363833.2| | BX363833 Homo sapiens B CELLS (RAMOS CELL LINE) COT 25-NORMALIZED Homo sapiens cDNA clone CS0DL006YE03 3-PRIME | 98,G/A/C/T | NO |
Table 9: SNPs from the secis element of the selenoprotein O.
All of these sequences can form a SecIS element.
There are two sequences (gi|46304542|emb|bx354948.2| and gi|46304039|emb|bx363833.2|) that
not have their corresponding image. We know that the SecIS is possible but we do not know
if there is an aberrant conformation.
The four sequences that contain only one SNP mentioned above, appear know with its images.
Therefore we think that only one SNP can alter the SecIS structure, but when we have more
than one SNP, the structure is viable due to the fact that the different changes make up for each
other.
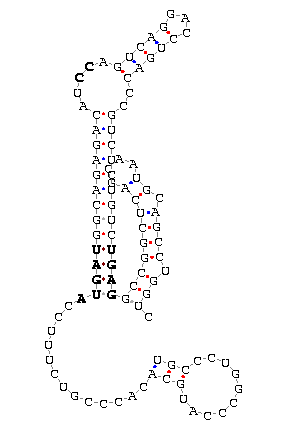 |
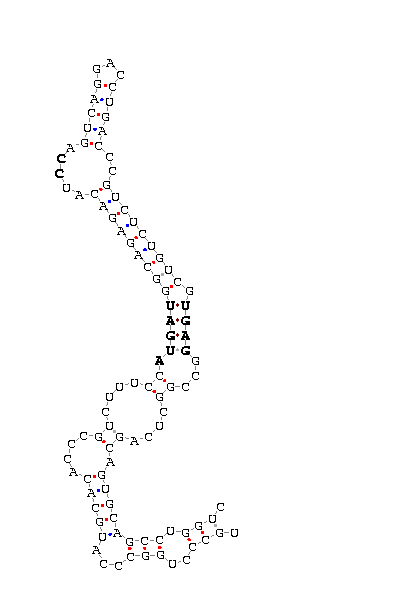 |
gi|46554667|emb|bx354872.2|
4c 89a 90t
| gi|46307094|emb|bx349459.2|
70- 73g
|
Figure 14: Secis structure from Sel O.
In this table we show two different cases of SecIS structures from the selenoprotein O.
The first one, corresponding to the identifier
gi|46554667|emb|bx354872.2| 4c 89a 90t, shows a strange SecIS structure. We do not know if it is due to
the program, that shows the real structure that will adopt the sequence in the cell, or that
this conformation is one of the multiple shapes that the sequences can adopt.
The second one corresponds to the identifier gi|46307094|emb|bx349459.2|70- 73g. Now the
SecIS structure seems to be correct but if we observe accurately the different matches, we
can see there is one nucleotide ("G" near the important motif of the SecIS structure)
that does not match with any nucleotide.
We can expect that the SecIS structure can not join SBP2, so the selenocysteine will not be
introduced into the peptid sequence.
Due to these facts, these two cases could be involved in neuroblastoma or some different diseases.
Relation between SecIS sequence and diseases:
As we have mentioned, some of the ests obtained in the blast hits
are related to neuroblastoma. This fact points to a possible relation between SNPs in the selenoprotein O
and this kind of disease. We wanted to know much about this possible relation so we
have been looking for information in many databases, but we have not found any article or review
explaining the possible relation between these two facts.