It is a medium-sized snake (up to 120cm). It has 17-21 rows of smooth and imbricate scales. Its head is slightly triangular and not distinct from neck and upper head is blackish and distinctly marked with a broad band of creamy or light cyan anterior to eyes and extending to sides of head. The body is elongated and rounded and slightly laterally compressed as its tail.
Laticauda laticaudata depends on the shore of coral islets for digestion, reproduction, skin sloughing and resting after foraging at sea.
The venomous sea snake Laticauda laticaudata is distributed in waters from the Bay of Bengal and the seas of the south Japan to the coast of Australia and islands of Oceania. It is commonly found on coral reefs and rocky areas along the sea cost.
Selenium (Se) is an essential micronutrient of several major metabolic pathways. Although Se can meet important physiological functions, there is a concentration limit between the beneficial effects and the toxic action on living organisms. In particular, selenium deficiency is associated with a wide variety of diseases, such as reduced antioxidant protection and cardiovascular disorders (Yang R et al., 2017). Contrarily, an accumulation of Se may cause an intoxication.
The biological effects of Se has been linked to selenium-containing proteins which refers to those involved in the composition of the protein with Se atom.
Selenoproteins exist in the three domains of live (archaea, bacteria and eukaryotes). These proteins include at least one selenocysteine (Sec, U, Se-Cys) amino acid residue, which is considered the 21st amino acid. Selenocysteine is similar to cysteine amino acid but differs by the presence of a Se atom instead of a sulfur (S).
Most Se in animal exists in selenoprotein form. Se is cotranslationally incorporated into the polypeptide chain as part of the amino acid Sec. Sec is a newly recognidez 21st coded amino acid which incorporation into the protein is mediated by codon UGA, which is also a stop codon.
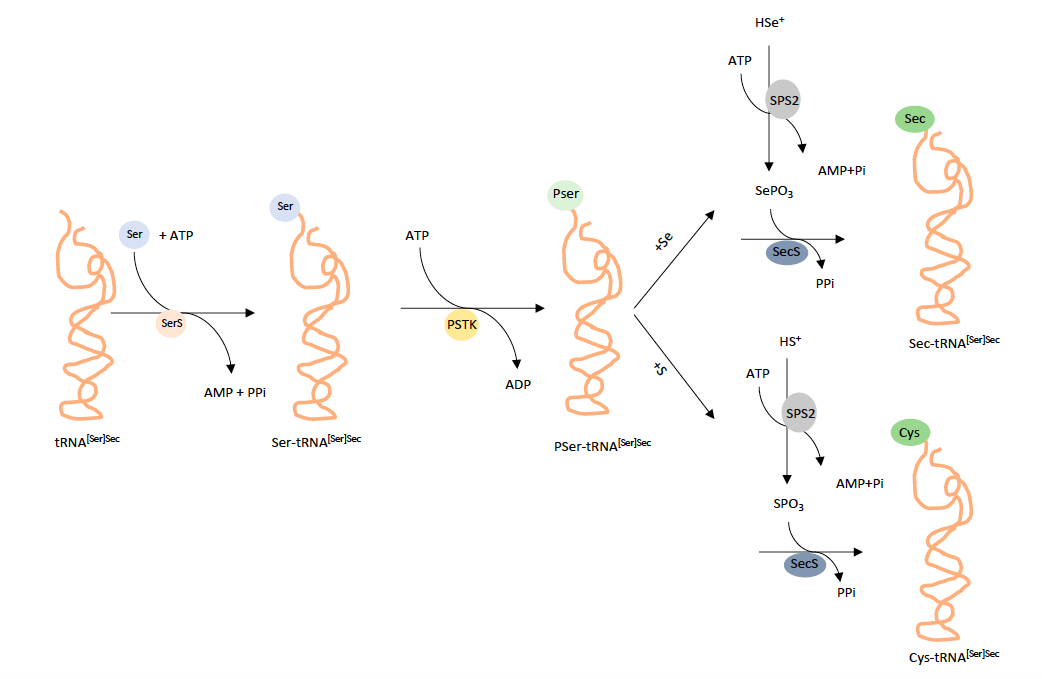
In the biosynthesis of Sec, tRNA[Ser]Sec is aminoacylated with serine by SerS and the seryl moiety is then converted to O-phosphoseryl-tRNA[Ser]Sec by PSTK. SecS acts on O-phosphoseryl-tRNA[Ser]Sec to hydrolyze the phosphate group forming the acceptor molecule, likely dehydroalanyl-tRNA[Ser]Sec, which in turn accepts the active Se donor, selenophosphate, synthesidezed by SPS2. SecS then converts the active acceptor to Sec, completing the biosynthesis of Sec (Turanov AA et al., 2011).
The mechanisms responsible for designating UGA as a Sec codon instead of termination involve a stem-loop structure in the 3'UTR of selenoprotein mRNA in eukaryotes known as SECIS (Sec Insertion Sequence) element and a SECIS binding protein, designated SBP2. SBP2 binds to the SECIS element and forms a complex with the specific elongation factor for Sec tRNA[Ser]Sec, EFSec, for incorporation of Sec into protein in response to the UGA Sec codon. Because the synthesis of some selenoproteins is terminated by a UGA stop signal, the Sec insertion machinery has the capability to distinguishing between the UGA for amino acid incorporation and the UGA for termination. The distance between the Sec codon and the SECIS element plays an essential role in this distinction (Turanov AA et al., 2011).
The prediction and annotation of selenoprotein genes is a difficult task due to the lack of an effective method for distinguishing the dual function of the UGA codon in the open reading frame as it acts as a Sec residue in the case of selenoproteins and traditionally, as a stop codon. Even known, selenoproteins are often misannotated in genome databases (Hua Chen, 2012).
Since missanotation invariably involves the deletion of the region in the protein sequence including Sec-UGA, misprediction in the case of selenoproteins have the additional negative effect (Mariotti M, 2010).
Application of bioinformatics methods to the identification of selenoproteins in newly sequenced species has been advantageous to study the selenoproteins.
The pipeline is a general homology-based gene finder program with specific features. (Mariotti M, 2010). In addition, the methods rely on the prediction of SECIS elements in coordination with the prediction of genes in which the strong codon bias characteristic of protein coding regions extends beyond a UGA codon interrupting an open reading frame (Liang Jiang, 2012).