Introduction
Selenium
The micronutrient selenium (Se) develops an essential function in the organism of living beings, as it is involved in the modulation of oxidative stress, redox signalling, immunity and protein folding (2). Se deficiency is directly related to different human diseases as Kashin-Beck disease, Myxedematous Endemic Cretinism, Keshan disease and male infertility (10). On the other hand, high levels of Se clearly show toxicity. These facts suggest that this micronutrient is strictly concentration dependent and for that reason has to be maintained on a certain narrow window in the organism in order to be beneficial for human health (4). Se binds to the active site of proteins forming selenocysteines and thus can produce its biological effects. The group of proteins that contain selenocysteine (Sec), also known as the 21th amino acid, are called selenoproteins (3, 5).
Selenoproteins biosynthesis
In the translational process, Sec is inserted to the selenoprotein encoded by a UGA stop codon, but the presence of an RNA stem-loop structure located in the 3’ untranslated region of eukaryotic selenoprotein messages, the Selenocysteine Insertion Sequence (SECIS), allows the insertion of a Sec instead of what would normally be a stop signal (5,6). The location of the SECIS element is not a functional necessity, but an evolutionary adaptation to enable a more efficient synthesis of selenoproteins (5). More elements are involved to this translational recoding event: a special Sec-tRNA, a specific elongation factor (EFSec) and an RNA-binding protein involved in the essential and critical interaction of all the machinery with the SECIS element of the selenoprotein mRNA, the SECIS-binding protein 2 (SBP2). Several other regulatory proteins are part of this complex cellular network that controls this recoding process, for example those involved in Sec-tRNA synthesis and selenocysteine recyclation (3,5,6).
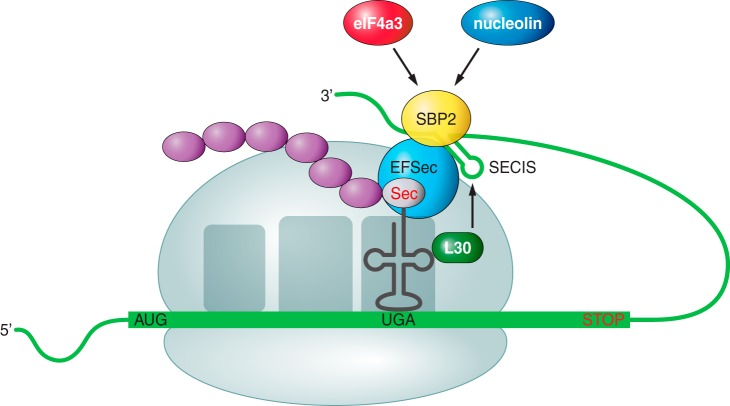
Recoding translational process. SBP2 mediates the interaction of the selenoprotein mRNA (by its SECIS element) with the translation machinery, allowing the insertion of a Sec encoded by a UGA codon. Other elements of the machinery are shown in the picture (7).
Human selenoproteome
The human selenoproteome, the set of selenoproteins in the genome, consists of 25 known selenoprotein genes. Glutathione peroxidase (GPx) enzyme family are an important member of this group, where there are 5 Sec-containing GPxs: cytosolic GPx1, gastrointestinal GPx2, plasma GPx3 and GPx6, and phospholipid hydroperoxide GPx4. Other group is the theoredoxin reductase (TR), which includes the cytosolic TR1, the mitochondrial TR3 and thioreduxin/glutathione reductase TGR. There are also 3 iodothyronine deiodinase (DI) selenoproteins. Other selenoproteins are SPS2, that participates in Sec biosynthesis by providing a selenium donor compound, and Selenoprotein W, the smallest mammalian selenoprotein. Finally, we have Selenoproteins V, Sep15, SelM, MsrB1, I, N, O, H, T, K, S and SelP (10).

Human selenoprotein genes. The selenoproteins are shown in alphabetical order. On the right, their relative lengths and Sec locations within the proteins, indicated by red vertical lines. (12)
Selenoproteins among evolution
Sec-containing proteins have been identified in all domains of life and have been conserved throughout evolution. Selenoproteomes are highly variable between species in eukaryotes. There is difference between for example plants and fungi, which do not have selenoproteins, with some fishes and algae, that have more than 30. In general, aquatic organisms generally have larger selenoproteomes than the terrestrial ones (13). However, closely related species show in general similarity between their selenoproteomes (10). Mammalian selenoproteomes contain generally 25 selenoproteins, but not all mammals have the same proteins in their genomes. The fact that the ancestral vertebrate is defined with 28 selenoproteins can suggest that there is a trend toward reduced use of selenoproteins in mammalian species. Selenoproteomes evolved along lineages through gene duplication, gene loss and replacement of Sec with Cysteine. (13)

Phylogenetic tree of changes in the vertebrates selenoproteome. The ancestral vertebrate selenoproteome is indicated in red, and the ones that are only found in vertebrates are underlined. In green, the creation of a new seleprotein. In grey, a loss. In blue, the replacement of Sec with Cys. No events of conversion of Cys to Sec were found. On the right, the number of selenoproteins predicted in each species. (13)
Genome prediction and annotation
The discovering of selenoproteomes in eukaryotic species helped to characterize the mechanisms and regulation of selenoproteins in different organisms. Indeed, it helped to understand the functions and metabolic pathways of Se, and therefore biosynthesis processes and functions of selenoproteins. Moreover, evolutionary trends of selenoproteome composition have been seen with these analysis, and that lead to a better understanding of the role of selenoproteins in human health and disease (10).
However, in the prior studies it has been always difficult to identify selenoproteins in databases due to the recognition of UGA codons as stop signals, as available computational tools lack the ability to correctly assign UGA function and selenoproteins were generally misannotated. To detect these sequences as selenoproteins, in specific to recognize the Sec-encoding UGA codon, new bioinformatics tools were needed. The development of these tools based on the recognition of SECIS elements, cysteine-containing homologs and coding nature of UGA codons, allowed the identification of the selenoproteomes in a lot of prokaryotic and eukaryotic species (6).
In previous literature, human selenoproteome could have been discovered by searching for already known selenoproteins from other species. Mouse and rat genomes were used as references, as these rodents were the first mammal species in which the complete genome was determined, because of the homology between rodents and humans. To prove the identity of selenoproteins obtained, analysis of SECIS elements should be performed. Rodents have 24 selenoproteins instead of 25, as this species do not have GPx6. Therefore, rodent selenoproteomes were found to be similar to human selenoproteome. Moreover, functionally important sequences such as SECIS elements remained conserved between them (10).
In our study, we aimed to characterize the selenoproteome of the rodent species Spermophilus dauricus by applying gene prediction and phylogenetic reconstruction methods. To determine it, we wanted to apply the same method of homology between species and for that reason the selenoproteome of Ictidomys tridecemlineatus, commonly known as squirrel, was used as reference. As the selenoproteome of squirrel appears registered in SelenoDB databases, the aim of the study was to associate by homology the sequences of these selenoproteins with sequences of the genome of Spermophilus dauricus in order to determine the selenoproteins on this species. In many cases, the squirrel genome was not useful due to different reasons (e.g. initial GAPs, no methionine in the beginning, truncated sequences, deficient annotation…). Therefore, we associated by homology the sequences of Homo sapiens selenoproteins with the Spermophilus dauricus ones.
To prove that the sequences obtained in Spermophilus dauricus by this method are selenoproteins, two other computational methods were used for the identification and analysis: SECISearch3, that predicts eukaryotic SECIS elements, and Seblastian, which predicts selenoprotein sequences encoded upstream of SECIS elements (11).
Spermophilus dauricus
Spermophilus Dauricus, commonly known as Daurian ground squirrel, is a species of rodent that belongs to Sciuridae family. It is present in dry plain steppes in Transbaikalia (Russia) and deserts in China, but is a considered a characteristic species of the north of Mongolia and the adjacent borders of Siberia (14). This rodent live on pastures, road borders, along railroads using burrows of marmots and daurian pika. Although it exists a possible habitat degradation due to the increasing numbers of livestock, this species is listed as Least Concern in Russia and China. Conservation only occurs in some protected areas in Mongolia (14). For more information related to geographic distribution, description and biology, check here