INTRODUCTION
Selenoproteins in living beings
In 1817 a Swedish chemist, Jacob Berzelius, discovered the element selenium (Se). He named it Selene, referring to the moon goddess in Greek. Se is an essential trace element in many organisms, including humans. It is known that this micronutrient supports a great number of important cellular functions. Human’s intake varies hugely worldwide ranging from deficient to toxic concentrations. Recommendations for selenium intake average 60 μg per day for men and 53 μg per day for women. [1] [2] [3].
The major form of Se in the cell is the 21st amino acid in the genetic code: selenocysteine (Sec) encoded by the UGA codon, which in many circumstances signals the end of translation. The groups of proteins that contain Sec as an integral part of their polypeptide chain are defined as selenoproteins and they are present in all lineages of life (prokarya, archaea and eukarya)[4] [5]. However, not all species have been reported to use them. For example, yeast and plants lost their selenoprotein insertion machinery during the evolution process. Moreover, it has been described that aquatic organisms count with more selenoproteins than terrestrial species [6].
There is a great number and a vast variety of selenoproteins, up until now, 45 selenoprotein families have been described in vertebrates [7].Nevertheless, all families together have never been declared to merge into single specie. Furthermore, researchers have confirmed a reduction on its number during evolution time, especially after terrestrial colonization. Likewise, the great number of selenoproteins in bony fish is due to the genome duplication that occurred within this clade [6].
Selenoproteins play an important role in human health. They have been reported to have biological functions in oxidoreductions, redox signaling, antioxidant defense, thyroid hormone metabolism, and immune responses. They thus possess a strong correlation with human diseases such as cancer, Keshan disease, virus infections, male infertility, and abnormalities in immune responses and thyroid hormone function [1][3][8].However, there are many things about selenoproteins that are still remained unknown and should be studied profoundly to have a better comprehension of the complexity surrounding the selenoproteome among the evolutionary tree.
Selenoproteins are a group of proteins that contain a Sec amino acid within the sequence that encodes for its gene. Sec is encoded by the UGA codon, which generally signals as a Stop codon. In the case of selenoproteins, this codon forms the Sec amino acid. The only difference between Sec and cysteine (Cys) residues is the presence of a Se atom instead of a sulfur radical. Some Sec mutate to Cys but keep their functionality. Thus, some members of selenoprotein families are analogs containing Cys [1][2] [6].
The mechanism by which selenoproteins are synthesized has been thoroughly described. Translation of selenoproteins is similar to generalized protein translation in that it consists of three main steps: initiation, elongation, and termination. But the special feature of selenoprotein translation lies in the UGA codon that, as it has been mentioned above, it experiences a duality phenomenon as it can code for a Stop codon or for a Sec-insertion codon [6][9].
Selenoproteins contain the named SECIS elements (SElenoCysteine Insertion Sequence), 60-nucleotide-length stem-loops that residue in the 3’-UTR of eukaryotic selenoprotein mRNA. The mechanism by which the UGA codon is recodified is mediated by the SECIS sequence [6] [7].The way that UGA Cys and Sec codons are distinguished resides in their position within the selenoprotein mRNA and in the accessibility of the SECIS element in the 3′-UTR. As long as the SECIS is buried in the 3′-UTR and thus unavailable, UGA codons are read as Cys. A ribosome approaching a UGA Sec codon triggers unfolding of the 3′-UTR, leading to unmasking of the SECIS and decoding of UGA Sec [7].
The UGA codon duality is circumvented by the presence of conserved cis- and trans- acting elements dedicated to the decoding of UGA as a Sec residue[10][11][12] The basic cis- acting elements for the synthesis of a selenoprotein is the SECIS element, as well as the in-frame UGA codon. On the other hand, the basic trans- acting elements are SPS1 (selenophosphate synthetase 1), SPS2 (selenophosphate synthetase 2), SecS (selenocysteine synthase), Pstk (O-phosphoryl-tRNASec kinase), eEFsec (eukaryotic elongation factor, selenocysteine-tRNA-specific), SBP2 (SECIS binding protein 2) and SeCys/tRNA[Ser]Sec (selenocysteyl-tRNA[Ser]Sec). All these factors are globally known as the selenoprotein translation machinery [5][10].
Interestingly, Sec is the only amino acid synthesized directly on its transfer RNA (tRNA[Ser]Sec), and is not produced from a Cys but from a serine (Ser). tRNA[Ser]Sec is initially aminoacylated with serine by the SerRS and then phosphorylated by Pstk, which will allow next reaction. SPS2 prepares the Se to be incorporated and SecS binds the atom to the Ser, completing the synthesis of the tRNA. This tRNA is not recognized by usual elongation factors, and is instead bound to the specific factor eEFSec [5][10]. Secp43 (tRNA selenocysteine 1 associated protein 1) is also involved in the synthesis of the tRNA[Ser]Sec and selenoproteins[7][13].
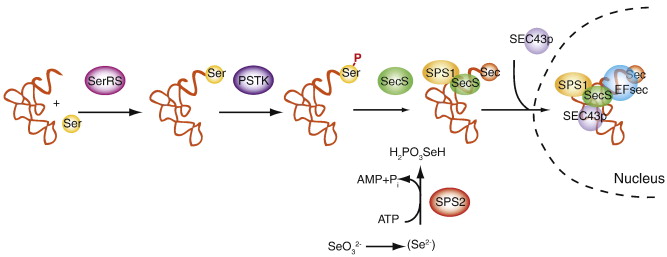
After the trans- acting elements have participated in the tRNA synthesis, the SECIS element acts in Cys to recode the UGA codon. SECIS recruits SPS2, which will in turn recruit and bind eEFSec. Finally, to achieve the incorporation of the Sec, the protein has to select the tRNA[Ser]Sec, which is taken to the UGA codon. From this point, the decoding process is able to continue in order to complete the whole selenoprotein translation [7].
We can distinguish two major groups of selenoproteins depending on their function. The first group is formed by housekeeping genes and the second group contains stress activated proteins. A description of the several selenoproteins families is summarized in the following table [6][11]:
| Name | Description |
| GpX | Glutathione peroxidase enzymes were the first to be characterized. They represent a major class of functionally important selenoproteins. Four have been described: classical GPx1, gastrointestinal GPx2, plasma GPx3 and phospholipid hydroperoxide GPx4 |
| TR | Thioredoxin reductase (TR) is a seleno-cysteine containing enzyme which catalyzes the NADPH dependent reduction of thioredoxin and therefore plays a regulatory role in its metabolic activity |
| ID | Iodothyronine deiodinase enzymes catalyze the 5'5-mono-deiodination of the pro-hormone thyroxin (T4) to the active thyroid hormone 3,3'5-triiodothyronine (T3) |
| SPS | The function of selenophosphate synthetase (SPS) is to generate the Se donor compound (selenophosphate) necessary for Sec biosynthesis |
| Sel15 | The specific function is not known. Sel15 levels differentially respond to selenium supplementation. Studies in mouse suggest that this selenoprotein may have redox function and may be involved in the quality control of protein folding |
| SelH | Selenoprotein H (Sel H) is a thioredoxin fold-like protein that contains a conserved Cys-X-X-Sec motif (X is any amino acid). Its expression is widely distributed throughout a variety of tissues and relatively high in early stages of embryonic development |
| SelI | A recent study has proposed that Selenoprotein I (Sel I, hEPT1) is involved in phospholipid synthesis as it is ubiquitously expressed in multiple tissues |
| SelK | Selenoprotein K is a small protein localized in the endoplasmic reticulum (ER) membrane. Some evidence suggests it is associated with the plasma membrane |
| SelM and Sep15 | Selenoprotein M (Sel M) and Sep15 are 15 kDa proteins that share 31% sequence identity and are localized in the ER. A link between Sep15 and cancer has been made by several in vitro studies |
| SelN | It is similar to Sel K and Sel S. SelN is a transmembrane protein localized in the ER membrane. Evidence suggests that Sel N serves to regulate calcium mobilization required for normal muscle development and differentiation. It remains to be seen whether Sel N regulates calcium mobilization in other tissues or under particular conditions, or if its role in the ER membrane is different in muscle versus other tissues |
| SelO | There is no information regarding its tissue distribution, subcellular location, or physiological role |
| SelP | Selenoprotein P (SelP) is a unique member of the selenoprotein family that has multiple Sec residues in a single protein, this suggests that this particular selenoprotein plays an important role in the Se transport. Recently, some studies suggest a relationship between SelP and Alzheimer’s disease |
| SelR | Sel R is part of the methionine sulfoxide reductase (Msr) family of proteins and is widely distributed throughout different tissues, although highest levels are found in liver and kidney. Suggested roles for Sel R in humans include protection from neurodegeneration, lens cell viability, and oxidative damage during aging |
| SelS | Selenoprotein S is a transmembrane protein located in the ER and plasma membranes and is widely expressed in a variety of tissues. It has been suggested to participate in the removal of mis-folded proteins from the ER lumen for degradation, to protect cells from oxidative damage and ER stress-induced apoptosis. |
| SelT | Selenoprotein T (SelT) is ubiquitously expressed throughout embryonic development, and it is most likely localized in the ER through a hydrophobic domain. The expression of SelT is proposed to be similar to those selenoproteins involved in stress-related phenomena |
| SelV | Expression of this selenoprotein appears to be restricted to testes, but its function in this tissue is unknown |
| SelW | Selenoprotein W is a small selenoprotein that contains a Cys-X-X-Sec motif. Protein and mRNA expression correlate in humans and are widely distributed throughout a variety of tissues |
| Name | Description |
| eEFsec | Eukaryotic elongation factor (eEFSec) |
| MsrA | Methionine Sulfoxide Reductase A is an enzyme that catalyses the reaction from methionine sulfoxide to methionine. This process also forms part of oxidative stress mechanisms. |
| pstk | Forms part of the machinery that allows selenoprotein synthesis. Catalyses the reaction from seryl-tRNA Ser (sec) to the phosphoryl form using a molecule of ATP. |
| SBP | Secis Binding Protein recognizes the SECIS element and recruits eEFSec and other factors in order to recode the UGA codon. |
| secp43 | Selenocysteine associated protein. Has two binding domains that allow it to recognize RNA motifs. It forms a complex with tRNAsec and participates in the synthesis of selenocysteine as well as the regulation of tRNAsec methylated levels. It also has a stabilizing role in the complex eEFsec - SBP2 -tRNAsec. |
| SecS | SecS binds the selenophosphate molecule to the Serine carried by the seryl-RNA. |
| SPS | The function of selenophosphate synthetase (SPS) is to generate the Se donor compound (selenophosphate) necessary for Sec biosynthesis. |
The next-generation sequencing technologies offer novel and rapid ways for genome-wide characterisation. This technology is also an available and useful tool for analyzing the selenoproteome because there are still many genomes that remain unstudied for selenoproteins.
he figure represents the evolution of the vertebrate selenoproteome. By observing the selenoproteome in vertebrates we can predict the phylogenetic tree points where duplications have occurred (indicated in green), if there has been a loss of a particular selenoprotein (indicated in grey), or whether there has been a replacement of Sec with Cys (indicated in blue)[6].
Selenoproteins are a useful tool to study evolutionary processes. If we look at the phylogenetic tree of the different families of selenoproteins in vertebrates we can see that the specie we are studying, Thunnus orientalis, does not figure in the classification of bony fishes, so to predict the different selenoproteins in this genome we will base our search on other species that are evolutionarily related to Thunnus orientalis such as the Zebrafish.

As our study is based in Thunnus orientalis, we are especially interested in the evolution of selenoproteins that involve fishes. Aquatic organisms have larger selenoproteomes in comparison with terrestrial organims. The vertebrate specie that contains the maximum number of selenoproteins is the zebra fish, another reason why we have selected this specie to compare it with the one we are interested. It has also been described that several selenoprotein genes are duplicated in certain lineages of bony fishes. Several selenoproteins were lost when colonization of the terrestrial environment happened, so some fishes have certain selenoproteins, such as SelL and SelJ that are not found in other vertebrate species. Other selenoproteins like Fep15 are identified in bony fishes, in cartilaginous fishes and its homologue as Cys is also found in the frog, so the conversion of Sec to Cys, which is not functionally equivalent, contributes in the reduction of the genome [6].
Thunnus orientalis
| Kingdom | Animalia |
| Phylum | Chordata |
| Subphylum | Vertebrata |
| Infraphylum | Gnathostomata |
| Superclass | Osteichtyes |
| Class | Actinopterygii |
| Order | Paciformes |
| Family | Scombridae |
| Genus | Thunnus |
| Subgenus | Thunnus |
| Species | Thunnus orientalis |
The pacific bluefin Tuna, known as Thunnus orientalis, is a migratory pelagic fish capable of swimming long distances at high speed. Its main habitat is the North Pacific Ocean, ranging from the East Asian coast to the western coast of North America.
A rare trait that differences bluefin tunas amongst other fishes is that they are warm-blooded, being able to adjust their body temperature which is why they are well adapted to cool ocean waters.
Their main source of energy includes small squids and fishes, sessile animals, red crabs and krill. They reach maturity at about 5 years of age, and spawning occurs from April to August, but the exact timing depends on the region.
The bluefin tuna species are one of the most important fishes regarding economically and culturally aspects in many parts of the world, reason why they are in steep decline due to overfishing.