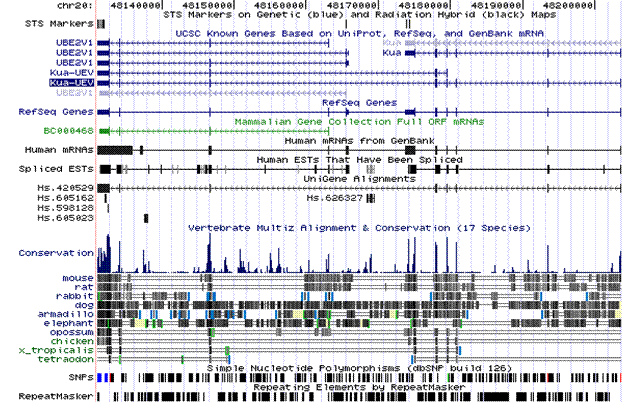
Fig.1 On the top part there is a representation of the transcripts of the two genes as well as the fusion of the genes. In the mid-table there is the conservation of the hybrid gene.
The results will be studied separatedly. Click the following links in order to see the results for each gene:
KUA-UBE2V1 UBE2V1 KUA
CONCLUSION
The hybrid gene Kua-UEV results from the fusion of two adjacent genes, Kua and UBE2V1, located in
chromosome 20q13.2, in strand minus.
These genes can either codify for two independent proteins or for the fusion protein Kua-UEV which mantains the function
of UEV but in a different cellular location.
UBE2V1 is a variant of the subfamily of ubiquitin-conjugating E2 enzymes, wich encodes for inactive E2 proteins.
This gene has five transcript variants generated by alternative splicing that codify for four isoforms. The resulting
proteins have between 103 and 170 aa, and are located in the nucleus.
Besides, the upstream gene Kua is a newly discovered pseudogene which only has one transcript that encodes for a 267aa
long protein and for whom there is no information about its function.
The fusion of both genes generates the variant Kua-UEV where domain B comes from UBE2V1 and the A domain comes from Kua.
This gene has two transcripts and two isoforms, one of 221aa and the other of 370aa. These proteins are known to mantain
the UBE2V1 function but a cytosolic localization.
Because the sequence of aminoacids is the same in all the variants, we asume that there is no reading-frameshift in any
of the genes' transcripts.
The two genes Kua and UBE2V1 have been well conserved throughout evolution because of a positive selection pressure.
There is a high conservation in those species that are more phylogenetically similar, and less in those that are more different.
According to the expression, the three genes (Kua, UEV and the fusion Kua-UEV) are observed to be highly expressed in blood cells and
immunitary cells as well as in the salivary glands and the respiratory system (like lungs or bronchial epithelium). There is
evidence that the three are underexpressed in brain structures like the amygdala or the caudate nucleus. However, there are organs such as
pancreas or ovaries where the expression can differ depending on the gene.
Because the gene Kua has no known function, and the hybrid Kua-UEV has the same function as UEV but in a different localization
we should only mention the function of UEV. The protein encoded by this gene is located in the nucleus and regulates noncanonical elongation of ubiquitin chains, a variant of polyubiquitination.
In mammalian cells, UEV proteins can modulate c-FOS transcription and the G2-M transition of the cell cycle. The overexpression of UEV1 in human colon cancer cells induces the accumulation of cells in G2-M and poliploidy, apoptosis, and inhibition of cell differentiation.
Regarding characterization of the genes' promoter regions, first of all we can conclude that of
the two methods used, the best one is PROMO server, because the bioinformatics are much more accurate.
Fusion Kua-UEV and Kua have the same promoter region, and thus the same TFs.
The coincident TFs between the two methods have been: NF-AT1, NF-kappaB and RXR-alpha for Kua-UEV and Kua and
NF-AT1, NF-kappaB, YY1, RXR-alpha, AhR and HNF-4 for UBE2V1.
Finally, we can conclude that biology is not as simple as it seems at first sight -one gene, one protein-, since gene
products can also be expressed as fused proteins, giving more complex functions to cells. In our subject of study
(Kua-UEV fusion) in order to obtain more information about the genes, more research on Kua should be carried.
REFERENCES
- Thomson TM, Lozano JJ, Loukili N, Carrio R, Serras F, Cormand B, Valeri M, Diaz VM,
Abril J, Burset M, Merino J, Macaya A, Corominas M, Guigo R. Fusion of the human gene for the polyubiquitination
coeffector UEV1 with Kua, a newly identified gene. Genome Res 2000 10: 1743-1756
-
Hofmann RM, Pickart CM. MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin
chains for DNA repair.Cell. 1999 Mar 5;96(5):645-53.
- Sancho E, Vila MR, Sanchez-Pulido LRole of UEV-1, an inactive variant of the E2 ubiquitin-conjugating enzymes, in in vitro differentiation and cell cycle
behavior of HT-29-M6 intestinal mucosecretory cells.Mol Cell Biol. 1998 Jan;18(1):576-89
Databases and Software
- NCBI: http://www.ncbi.nih.gov/
- Ensembl: http://www.ensembl.org/
- Clustalw: http://www.ebi.ac.uk/clustalw/
- UCSC: http://genome.ucsc.edu/
- PROMO 3.0: PROMO homepage