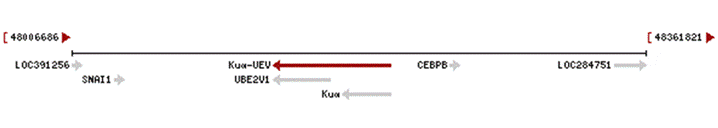
Co-transcription of the gene UBE2V1 (Ubiquitin-conjugating E2 enzyme variant 1) with the neighboring upstream gene Kua generates a rare transcript (Kua-UEV), which encodes a fusion protein comprised of sequence sharing identity with each individual gene product. Kua-UEV is a two-domain protein, where domain B comes from Kua, and confers the hybrid protein its cytoplasmic localization. This results in the fusion protein Ubiquitin-conjugating E2 enzyme variant Kua. The genes are located in chromosome 20q13.2. In strand minus.

Fig1. Representation of the fusion gene Kua-UEV in chromosome 20. From NCBI.
The gene encodes for two different transcripts due to alternative splicing:
Transcript variant 1: longer isoform
Transcript variant 2: shorter isoform
Different isoforms are a result of the alternative splicing process. In all alternative isoforms generated from this gene the reading frame is mantained, since the resulting protein has the same aminoacid sequence in all the isoforms.
List of orthologues from NCBI:
| IMAGE | SPECIE | GENE | PROTEIN
HOMOLOGY(%) |
DNA
HOMOLOGY(%) |
 |
P.troglodytes | LOC458329 | 99.7% | 99.8% |
 |
P.falciparum | MAL3P2.20 | 54.8% | 49.3% |
 |
O.sativa | OSJNBb003 | 50.4% | 55.9% |
 |
A.thaliana | AT1G23260 | 54.5% | 56.5% |
In humans Kua and UBE2V1 are located contiguously in the same chromosome. Meanwhile, in other species which are very separated phylogenetically from humans (like D. melanoganster and C.elegans) the gene for Kua is unlinked to the gene for the corresponding UEV protein, and Kua and UEV are always expressed as separate proteins. However, there is certain homology in primates such as P. troglodytes.
We have also found orthologues in plasmodium and two plants. This is quite abnormal, because considering that the fusion has probably occured in a recent evolution stage we should have not been able to find the hybrid gene in such species. Therefore, to explain this controversy (if the orthology data is correct, and the co-expresion does really exist in these species) we have hypothezised that the fusion could have also occured in these plants and plasmodium in a parallel way to that in humans. Because plants have a high rate of genomic duplications, this could increase the probability of the fusion between two different genes in different loci. Refered to plasmodium, because it is a parasit with a small genome, it is possible for the genes to be concentrated in the same chromosome, and thereby, near enough to co-express.
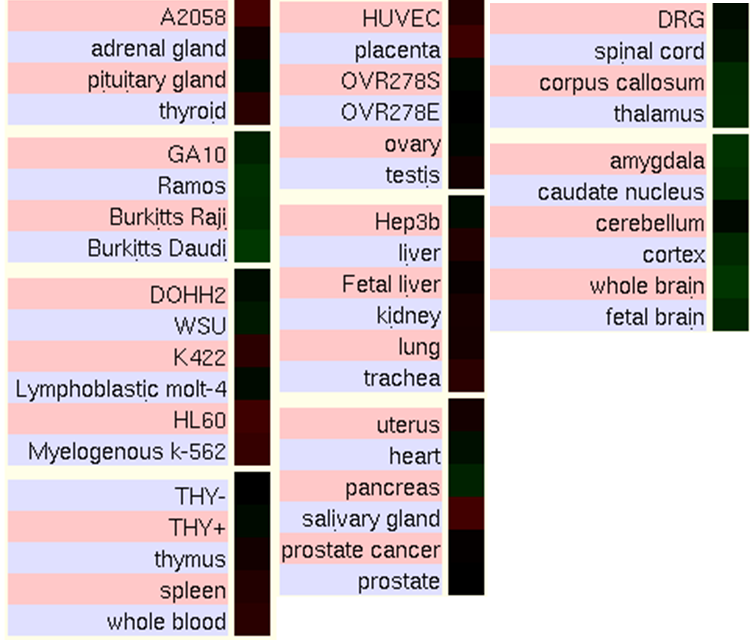
In order to characterize the gene expression in human cell types and tissues, some miroarray chips from UCSC Genome Browser have been analyzed. The figures show the ratios of two of these experiments, where red colour indicates the gene is highly expressed in the tissue and green that it is underexpressed.
Normal Human Tissue cDNA Microarrays
GNF Expression Atlas 2 Data from U133A and GNF1H Chips
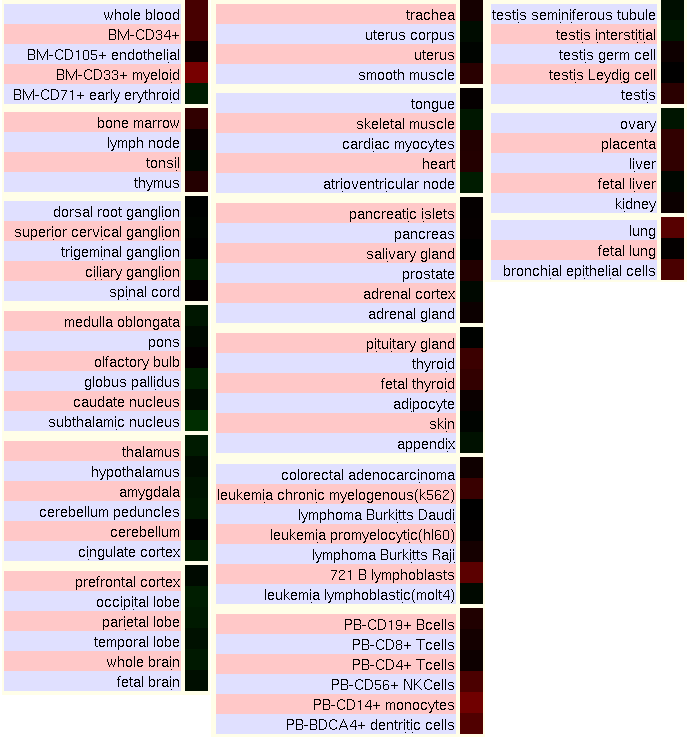
According to the data from the expression tables from UCSC, Kua-UBE2V1 is quite homogenicaly expressed. However, we can highlight that it is highly expressed in immune and blood cells, salivary gland, and the respiratory system like the trachea and lungs. It is detected at low levels mainly in pancreas, brain, bone and ovaries.
Comparing the expression of the hybrid protein with the expression of the two individual genes, it can be seen that tissues where there is over- or underexpression of Kua-UEV also have the same levels of expression either of UBE2V1 or of Kua. Because the function of the hybrid protein and the individual proteins is similar, it is logical to observe this correlation.
| MOLECULAR FUNCTION | |
| Ubiquitin-protein ligase activity GO:0004842 |
Catalysis of the reaction: ATP + ubiquitin + protein lysine = AMP + diphosphate + protein N-ubiquityllysine |
| Transcriptional activator activity GO:0016563 |
Any transcription regulator activity required for initiation or upregulation of transcription |
| BIOLOGICAL PROCESS | |
| Protein modification GO:0006464 |
The covalent alteration of one or more amino acids occurring in a protein. This includes pre-translational, co-translational and post-translational modifications |
| Ubiquitin cycle GO:0006512 |
The cyclical process by which one or more ubiquitin moieties are added to (ubiquitination) and removed from (deubiquitination) a protein |
| Regulation of progression through cell cycle GO:0000074 |
Any process that modulates the rate or extent of progression through the cell cycle |
| Protein polyubiquitination GO:0000209 |
Addition of multiple ubiquitin moieties to a protein, forming a ubiquitin chain |
| Cell differentiation GO:0030154 |
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate |
| Regulation of DNA repair GO:0006282 |
Any process that modulates the frequency, rate or extent of DNA repair |
The Kua-UEV mRNA is an infrequent but naturally occurring co-transcribed product of the neighboring Kua and UBE2V1 genes. Two alternative transcripts encoding different isoforms have been described. The proteins produced by these transcripts have UEV1 B domains but the proteins are localized to the cytoplasm rather than to the nucleus. The significance of these co-transcribed mRNAs and the function of their protein products have not yet been determined.
The characterizations of the gene's promoter region and the transcription factors that are likely to bind to it has been done by two ways: with the program PROMO and with a PERL program developed by ourselves.
1.Perl program:
The folowing items are needed:
- The promoter sequence of the gene Kua-UEV
- Document TFs matrices, which contains 13 selected TFs matrices
- Perl program PERL PROGRAM(identificacio_de_lufs.pl), which has been developed by the authors.
The program has to be executed in Linux terminal by typing:
$ ./identificacio_de_lufs.pl humanfactors_selectedmatrices.txt sequenciapromotora_Kua-UEV.fa After running the program several times we show the obtained results in the following table, where the position in the promoter region the TF binds to and the score and p-value are noted:| Transcription factor | Position | Score | p-value |
| AP-1 | 818-824 | 4.40504 | 0.46 |
| AR | 299-305 | 5.93511 | 0.24 |
| c-Myc | 64-69 | 1.61938 | 0.77 |
| NF-AT1 | 96-102 | 1.47919 | 0.64 |
| NF-kappaB | 64-72 | 1.57878 | 0.83 |
| SRF | 64-72 | 1.40820 | 0.08 |
| YY1 | 285-290 | 6.66650 | 0.15 |
| RXR-alpha | 1123-1128 | 4.60764 | 0.86 |
| HIF-1 | 57-65 | 1.56189 | 0.75 |
| AhR | 207-213 | 7.04460 | 0.22 |
| PU.1 | 57-63 | 0.674430 | 0.8 |
| HNF-4 | 59-66 | 1.58026 | 0.61 |
| NRSF | 55-63 | 1.466530 | 0.45 |
We have reduced the list of TFs provided by PROMO by selecting the ones with smaller RE-equally values (using a 0,1 cutoff). RE-equally gives the number of expected occurrences of the match in a random sequence of the same length as the query sequence according to the dissimilarity index (15%) consifering equiprobability for the 4 nucleotides.
We can compare the results obtained with the two different methods, bearing in mind however that the program is just a very simple approach to the problem, and hence has a limited predictability.
The coincident TFs are the following: NF-AT1, NF-kappaB and RXR-alpha.