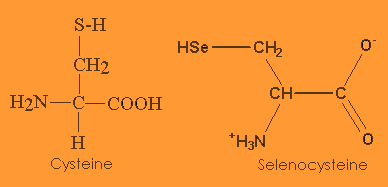
Selenocysteine, the 21st aminoacid (named sec or by the letter U), has a similar structure to cysteine, but with an atom of selenium taking the place of the usual sulfur.

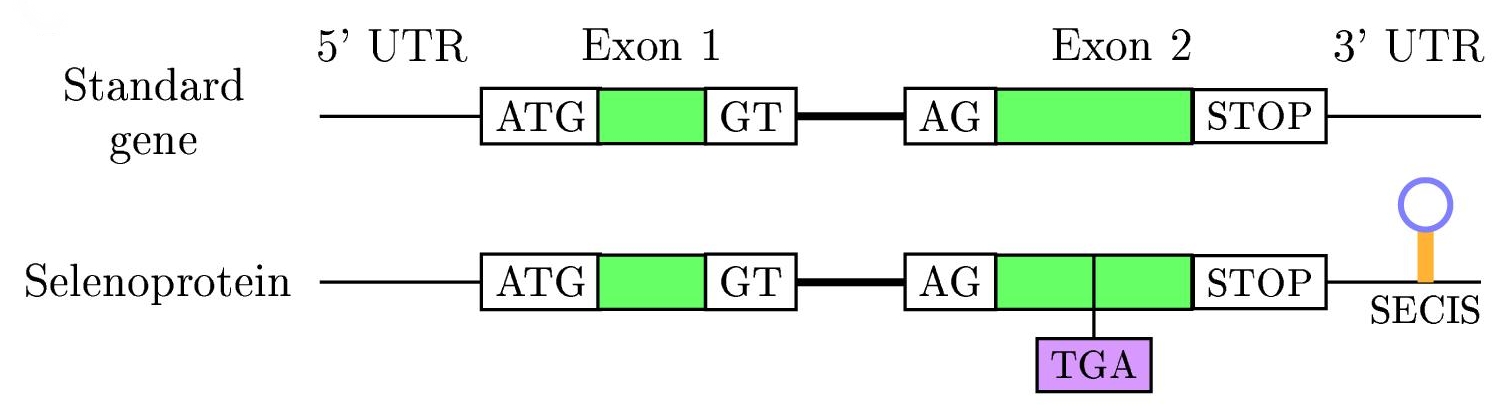
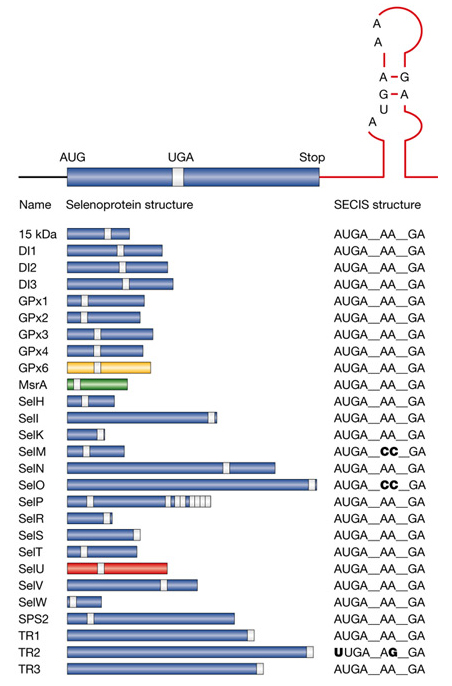
Figure 4: Selenoproteins in humans
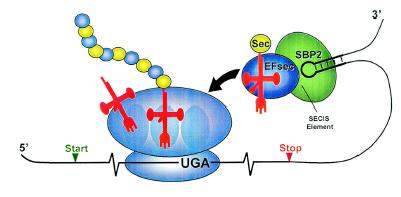
In all three domains, translation of UGA (usually codon stop) as selenocysteine requires distinct signals in the mRNA, which are SECIS elements: Selenocysteine Insertion Sequence.
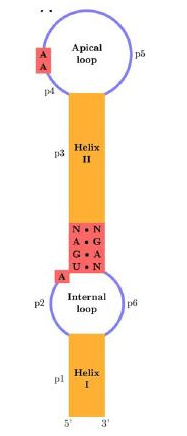
The SECIS element is defined by the fact that particular nucleotids tend to be at particular positions and because of the secondary structure base-pairing patterns, that causes a hairpin-like structure (consisting in a helix-loop-helix-loop, see right figure).
The eukaryotic SECIS element includes non-canonical A-G base pairs, which are uncommon in nature, but are critically important to correct SECIS element function. Although the eukaryotic, archaeal and eubacterial SECIS elements share a general hairpin structure, they are not alignable.
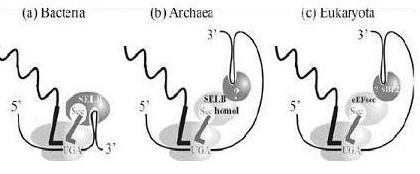
In eubacteria, the SECIS element is located immediately following the UGA codon within the reading frame for the selenoprotein. In archaea and in eukaryotes, the SECIS element is in the 3' UTR of the mRNA, and can direct multiple UGA codons to encode selenocysteine residues.
Like the other aminoacids used by cells, selenocysteine has a specialized tRNA, that differs from the other standards tRNAs by a long variable region arm. The selenocysteine tRNAs are initially charged with serine by seryl-tRNA ligase, but the resulting Ser-tRNA(Sec) is not used for translation because it is not recognised by the normal translation factor (EF-Tu in bacteria, EF1alpha in eukaryotes). The tRNA-bound seryl residue is converted to a selenocysteyl-residue by the pyridoxal phosphate-containing enzyme selenocysteine synthase. Finally, the resulting Sec-tRNA(Sec) is specifically bound to an alternative translational elongation factor (SelB or mSelB) which delivers it to the ribosomes translating mRNAs for selenoproteins. The specificity of this delivery mechanism is brought about by the presence of an extra protein domain (in bacterial SelB) or an extra subunit (SBP-2 for eukaryotic mSelB) which bind to the corresponding RNA secondary structures formed by the SecIS elements in selenoprotein mRNAs (see figure below).