SECIS
- The SECIS element (SECIS: selenocysteine insertion sequence) is a structural motif (pattern of nucleotides) that directs the cell to translate UGA codons as selenocysteines, not as stop codons.
- Secis elements are common to all selenoproteins. They are located in the 3'UTR in eukaryotes and archaea, while in the coding region in bacteria (after the UGA). Eukarya, Bacteria and Archaea SECIS elements differ substantially.
- The SECIS element appears defined by sequence characteristics, i.e. particular nucleotides tend to be at particular positions in it, and a characteristic secondary structure, that it is the result of base-pairing of complementary RNA nucleotides, and causes a hairpin-like structure.
- The eukaryotic SECIS element includes non-canonical A-G base pairs, which are uncommon.
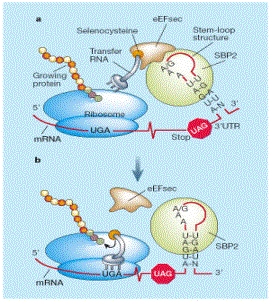 |
| Figure 1:Translation of mRNA requires a SECIS-element in case of selenocysteine. |
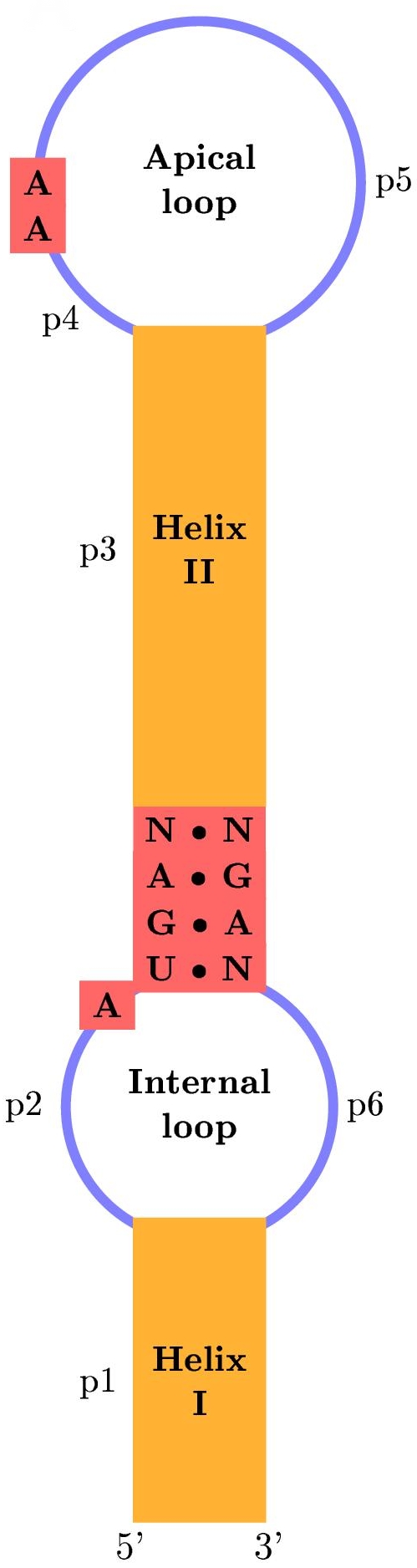 |
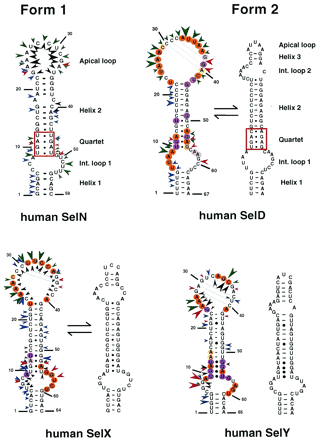 |
| (A) |
(B) |
This scheme shows the basic secis strucutral motif, with the critical positions marked in red (A), and some of the "real" secis elements found in 3'UTR mRNA human transcripts (B).
- Eukaryotic selenoproteins remain poorly characterized. The dual role of the UGA codon confounds the identification of novel selenoprotein genes. To achieve a reliable results in predicting selenoproteins within genomes has proven to be difficult.
- Nowadays, comparative genomics approaches rely on the genome-wide prediction of genes with in-frame TGA codons, and the subsequent comparison of predictions from different genomes, wherein conservation in regions flanking the TGA codon suggests selenocysteine coding function.
- Actually, there are many programs used to predict selenoproteins which are available through the Internet:
Geneid is used to predict selenoprotein genes in human genome (among others).
Secisearch allows you to find "posible" secis elements within a specific sequence.
Click the link to obtain more information about the secis elements of the selenoproteins under study:





