| | | | | |
genewise output
Score 201.81 bits over entire alignment
Scores as bits over a synchronous coding model
Warning: The bits scores is not probablistically correct for single seqs
See WWW help for more info
15-kDa 18 LLATVLQAVSAFGAEFSSEACRELGFSSNLLCSSCDLLGQFNLLQLDPD
L L +SA+GAE+SSEACRELGFSSNLLC+SCDLLG+F+L +L P+
LFPFALLQLSAYGAELSSEACRELGFSSNLLCTSCDLLGEFSLAKLQPN
SCAF14367 -205823 ctctgcccctgtgggcttggtagcgttaacctattgccggtacgaccca
ttctcttatccagcatccacggatgtcgattgccgattgatgtcataca
acgttccggagtccggaggccgggcccccggccgccgcagccccgcgcc
15-kDa 67 CRGCCQEEAQFETKK LYAGAILEVCG
C+ CCQ+EA +E +K LY GAILEVCG
CQQCCQQEALMEGRK LYPGAILEVCG
SCAF14367 -205676 tccttccggcaggcaGTAAGGC Intron 1 TAGctcggacggtg
gaaggaaacttagga<0-----[205631:20445-0>tacgcttatgg
cggctgggcggaacg gtcgctgggca
15-kDa 93 -KLGRFPQVQ FVRSDKPKLFRGL
KLGRFPQVQ FVRS+KPK+F+GL
XKLGRFPQVQ A:A[gct] FVRSEKPKMFKGL
SCAF14367 -204418 tacgatccgcGGTGAGCC Intron 2 TAGCTtgaagacaatagc
gatggtcata <1-----[204387:20418-1> ttggaacattagt
aagggccaca tcgtggaggcgtt
15-kDa 116 QIK YVRGSDPVLKLLDDNGNIAEELS
QIK YVRGSDPVLKLLDD GNIAEELS
QIK YVRGSDPVLKLLDDEGNIAEELS
SCAF14367 -204140 caaGTAAGAG Intron 3 CAGtgagtgcgcaccggggaagggca
ata<0-----[204131:20405-0>atggcacttattaaagatcaatg
gcg ccacactgcgtgccagcttggcc
15-kDa 142 ILKWNTDSVEEFLSEKLERI
ILKWNTDSVEEFLSEKL+RI
ILKWNTDSVEEFLSEKLDRI
SCAF14367 -203985 acataagagggtcagatgca
ttagacagtaattgaatagt
ccggcgccgggcgcgaatga
//
Gene 1
Gene 205823 203926
Exon 205823 205632 phase 0
Exon 204451 204388 phase 0
Exon 204181 204132 phase 1
Exon 204054 203926 phase 0
//
>SCAF14367.[205823:203924].sp.tr
LFPFALLQLSAYGAELSSEACRELGFSSNLLCTSCDLLGEFSLAKLQPNCQQCCQQEALM
EGRKLYPGAILEVCG*KLGRFPQVQAFVRSEKPKMFKGLQIKYVRGSDPVLKLLDDEGNI
AEELSILKWNTDSVEEFLSEKLDRI
//
>SCAF14367.[205823:203924].sp
CTATTCCCGTTTGCTCTCCTCCAGCTGTCAGCGTATGGCGCCGAGCTGTCATCGGAGGCC
TGCAGGGAGCTGGGCTTCTCCAGCAACCTGCTGTGCACCTCGTGCGACCTGCTCGGAGAG
TTCAGCCTCGCCAAGCTCCAGCCCAACTGCCAGCAGTGCTGTCAGCAGGAGGCCCTGATG
GAAGGACGCAAGCTGTATCCCGGGGCCATTCTGGAGGTGTGCGGATGAAAACTGGGGAGG
TTCCCCCAAGTCCAAGCTTTTGTCAGGAGTGAGAAGCCAAAGATGTTCAAGGGTCTTCAG
ATCAAGTACGTCAGAGGCTCAGACCCTGTGCTCAAGCTTCTGGACGACGAAGGGAACATT
GCTGAGGAGCTCAGCATCCTCAAGTGGAACACGGACAGCGTGGAGGAGTTCCTGAGCGAG
AAATTAGATCGGATA
//
SCAF14367 GeneWise match 205823 203926 201.81 - . SCAF14367-genewise-prediction-1
SCAF14367 GeneWise cds 205823 205632 0.00 - 0 SCAF14367-genewise-prediction-1
SCAF14367 GeneWise intron 205631 204452 0.00 - . SCAF14367-genewise-prediction-1
SCAF14367 GeneWise cds 204451 204388 0.00 - 0 SCAF14367-genewise-prediction-1
SCAF14367 GeneWise intron 204387 204182 0.00 - . SCAF14367-genewise-prediction-1
SCAF14367 GeneWise cds 204181 204132 0.00 - 2 SCAF14367-genewise-prediction-1
SCAF14367 GeneWise intron 204131 204055 0.00 - . SCAF14367-genewise-prediction-1
SCAF14367 GeneWise cds 204054 203926 0.00 - 0 SCAF14367-genewise-prediction-1

## gff-version 2 ## date Sun Feb 29 19:20:55 2004 ## source-version: geneid v 1.1 -- geneid@imim.es # Sequence SCAF14367 - Length = 444931 bps # Optimal Gene Structure. 67 genes. Score = 1668.568223 # Gene 1 (Reverse). 7 exons. 390 aa. Score = 31.771498 SCAF14367 geneid_v1.1 Terminal 2908 3090 6.34 - 0 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 3172 3338 6.32 - 2 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 3707 4098 4.89 - 1 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 5827 5999 6.47 - 0 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 6077 6243 7.88 - 2 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 6314 6398 -0.06 - 0 SCAF14367_1 SCAF14367 geneid_v1.1 First 7269 7271 -0.07 - 0 SCAF14367_1 # Gene 2 (Reverse). 5 exons. 203 aa. Score = 21.377901 SCAF14367 geneid_v1.1 Terminal 9582 9836 7.13 - 0 SCAF14367_2 SCAF14367 geneid_v1.1 Internal 10509 10675 9.24 - 2 SCAF14367_2 SCAF14367 geneid_v1.1 Internal 11010 11071 1.34 - 1 SCAF14367_2 SCAF14367 geneid_v1.1 Internal 11188 11292 1.80 - 1 SCAF14367_2 SCAF14367 geneid_v1.1 First 11496 11515 1.86 - 0 SCAF14367_2 # Gene 3 (Reverse). 1 exons. 67 aa. Score = 1.468603 SCAF14367 geneid_v1.1 Single 12189 12389 1.47 - 0 SCAF14367_3 # Gene 4 (Forward). 1 exons. 71 aa. Score = 4.075906 SCAF14367 geneid_v1.1 Single 16057 16269 4.08 + 0 SCAF14367_4 # Gene 5 (Reverse). 5 exons. 221 aa. Score = 2.085121 SCAF14367 geneid_v1.1 Terminal 17951 18066 1.57 - 2 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 18402 18479 0.32 - 2 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 22466 22556 -0.91 - 0 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 24219 24486 3.31 - 1 SCAF14367_5 SCAF14367 geneid_v1.1 First 25605 25714 -2.21 - 0 SCAF14367_5 # Gene 6 (Forward). 2 exons. 88 aa. Score = 4.024337 SCAF14367 geneid_v1.1 First 26527 26568 3.10 + 0 SCAF14367_6 SCAF14367 geneid_v1.1 Terminal 28117 28338 0.92 + 0 SCAF14367_6 # Gene 7 (Forward). 1 exons. 57 aa. Score = 1.928624 SCAF14367 geneid_v1.1 Single 31053 31223 1.93 + 0 SCAF14367_7 # Gene 8 (Reverse). 1 exons. 63 aa. Score = 0.996763 SCAF14367 geneid_v1.1 Single 33644 33832 1.00 - 0 SCAF14367_8 # Gene 9 (Forward). 8 exons. 753 aa. Score = 59.830281 SCAF14367 geneid_v1.1 First 35647 35906 1.59 + 0 SCAF14367_9 SCAF14367 geneid_v1.1 Internal 38840 39192 2.89 + 1 SCAF14367_9 SCAF14367 geneid_v1.1 Internal 41410 41633 5.69 + 2 SCAF14367_9 SCAF14367 geneid_v1.1 Internal 43215 43320 2.49 + 0 SCAF14367_9 SCAF14367 geneid_v1.1 Internal 43490 43726 8.49 + 2 SCAF14367_9 SCAF14367 geneid_v1.1 Internal 44585 44807 6.76 + 2 SCAF14367_9 SCAF14367 geneid_v1.1 Internal 45858 45918 2.70 + 1 SCAF14367_9 SCAF14367 geneid_v1.1 Terminal 46187 46981 29.22 + 0 SCAF14367_9 # Gene 10 (Forward). 1 exons. 28 aa. Score = 1.492212 SCAF14367 geneid_v1.1 Single 50672 50755 1.49 + 0 SCAF14367_10 # Gene 11 (Forward). 14 exons. 752 aa. Score = 31.808722 SCAF14367 geneid_v1.1 First 52362 52484 -1.23 + 0 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 52588 52634 2.28 + 0 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 52861 53002 4.10 + 1 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 53088 53273 4.06 + 0 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 53519 53609 0.54 + 0 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 54054 54112 -0.19 + 2 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 54194 54390 9.36 + 0 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 54463 54576 4.15 + 1 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 54686 54890 -0.29 + 1 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 55913 56144 1.68 + 0 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 56456 56670 0.88 + 2 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 56805 57020 2.32 + 0 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 57121 57299 -0.49 + 0 SCAF14367_11 SCAF14367 geneid_v1.1 Terminal 57952 58201 4.65 + 1 SCAF14367_11 # Gene 12 (Reverse). 1 exons. 108 aa. Score = 1.577283 SCAF14367 geneid_v1.1 Single 61083 61406 1.58 - 0 SCAF14367_12 # Gene 13 (Reverse). 1 exons. 55 aa. Score = 0.946988 SCAF14367 geneid_v1.1 Single 63647 63811 0.95 - 0 SCAF14367_13 # Gene 14 (Forward). 5 exons. 179 aa. Score = 23.144973 SCAF14367 geneid_v1.1 First 64853 64896 -1.07 + 0 SCAF14367_14 SCAF14367 geneid_v1.1 Internal 64977 65066 4.11 + 1 SCAF14367_14 SCAF14367 geneid_v1.1 Internal 65139 65200 3.60 + 1 SCAF14367_14 SCAF14367 geneid_v1.1 Internal 65279 65442 8.03 + 2 SCAF14367_14 SCAF14367 geneid_v1.1 Terminal 65521 65697 8.47 + 0 SCAF14367_14 # Gene 15 (Reverse). 1 exons. 83 aa. Score = 2.265268 SCAF14367 geneid_v1.1 Single 67176 67424 2.27 - 0 SCAF14367_15 # Gene 16 (Forward). 9 exons. 422 aa. Score = 34.941130 SCAF14367 geneid_v1.1 First 70356 70460 2.24 + 0 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 70899 70970 0.86 + 0 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 72110 72202 7.30 + 0 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 72277 72335 -2.05 + 0 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 72406 72514 8.24 + 1 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 72857 72887 -1.62 + 0 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 73109 73233 2.64 + 2 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 74613 74781 6.28 + 0 SCAF14367_16 SCAF14367 geneid_v1.1 Terminal 75195 75697 11.05 + 2 SCAF14367_16 # Gene 17 (Forward). 14 exons. 817 aa. Score = 55.697304 SCAF14367 geneid_v1.1 First 76502 76629 -1.20 + 0 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 79205 79442 10.09 + 1 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 79551 79698 9.06 + 0 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 79781 79883 -0.50 + 2 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 79999 80182 6.16 + 1 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 80274 80514 5.96 + 0 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 80787 80980 5.14 + 2 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 81065 81266 4.90 + 0 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 81349 81560 -0.26 + 2 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 81690 81817 6.19 + 0 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 81906 82124 5.42 + 1 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 82198 82432 4.21 + 1 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 82546 82695 0.21 + 0 SCAF14367_17 SCAF14367 geneid_v1.1 Terminal 85219 85287 0.31 + 0 SCAF14367_17 # Gene 18 (Forward). 13 exons. 517 aa. Score = 50.120702 SCAF14367 geneid_v1.1 First 86227 86482 11.15 + 0 SCAF14367_18 SCAF14367 geneid_v1.1 Internal 87166 87346 -0.55 + 2 SCAF14367_18 SCAF14367 geneid_v1.1 Internal 87663 87734 0.66 + 1 SCAF14367_18 SCAF14367 geneid_v1.1 Internal 88148 88325 10.42 + 1 SCAF14367_18 SCAF14367 geneid_v1.1 Internal 91676 91911 7.54 + 0 SCAF14367_18 SCAF14367 geneid_v1.1 Internal 92527 92560 -2.24 + 1 SCAF14367_18 SCAF14367 geneid_v1.1 Internal 92635 92721 3.47 + 0 SCAF14367_18 SCAF14367 geneid_v1.1 Internal 93254 93355 5.61 + 0 SCAF14367_18 SCAF14367 geneid_v1.1 Internal 93420 93512 5.59 + 0 SCAF14367_18 SCAF14367 geneid_v1.1 Internal 93587 93654 -0.58 + 0 SCAF14367_18 SCAF14367 geneid_v1.1 Internal 93724 93802 6.36 + 1 SCAF14367_18 SCAF14367 geneid_v1.1 Internal 93863 93945 1.16 + 0 SCAF14367_18 SCAF14367 geneid_v1.1 Terminal 94015 94096 1.53 + 1 SCAF14367_18 # Gene 19 (Reverse). 9 exons. 432 aa. Score = 27.528512 SCAF14367 geneid_v1.1 Terminal 95811 96115 5.52 - 2 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 96198 96325 3.13 - 1 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 96403 96530 1.03 - 0 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 96660 96754 2.04 - 2 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 96840 96921 3.12 - 0 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 97020 97171 5.11 - 2 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 97333 97506 2.19 - 2 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 97594 97723 1.96 - 0 SCAF14367_19 SCAF14367 geneid_v1.1 First 98123 98224 3.42 - 0 SCAF14367_19 # Gene 20 (Reverse). 15 exons. 744 aa. Score = 33.137352 SCAF14367 geneid_v1.1 Terminal 100808 101011 5.68 - 0 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 101105 101402 7.08 - 1 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 101482 101733 2.35 - 1 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 103651 103775 1.47 - 0 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 103894 104009 0.53 - 2 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 104087 104151 -0.02 - 1 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 104225 104427 5.59 - 0 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 104499 104631 2.17 - 1 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 104706 104861 0.37 - 1 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 104960 105071 1.05 - 2 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 105368 105419 -0.90 - 0 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 107800 108111 8.39 - 0 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 108424 108487 -1.36 - 1 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 110427 110448 -0.92 - 2 SCAF14367_20 SCAF14367 geneid_v1.1 First 110754 110871 1.66 - 0 SCAF14367_20 # Gene 21 (Reverse). 2 exons. 55 aa. Score = 0.796996 SCAF14367 geneid_v1.1 Terminal 118701 118747 -0.85 - 2 SCAF14367_21 SCAF14367 geneid_v1.1 First 118981 119098 1.65 - 0 SCAF14367_21 # Gene 22 (Forward). 1 exons. 59 aa. Score = 0.311036 SCAF14367 geneid_v1.1 Single 122733 122909 0.31 + 0 SCAF14367_22 # Gene 23 (Forward). 21 exons. 1201 aa. Score = 96.501152 SCAF14367 geneid_v1.1 First 124866 125034 0.92 + 0 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 128275 128373 4.04 + 2 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 129972 130092 3.62 + 2 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 130178 130226 3.45 + 1 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 130370 130552 6.97 + 0 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 130633 130749 4.96 + 0 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 130836 130978 7.73 + 0 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 131083 131259 6.63 + 1 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 131328 131402 2.84 + 1 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 131491 131661 3.22 + 1 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 131745 132008 10.36 + 1 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 136084 136303 2.97 + 1 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 138108 138459 6.32 + 0 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 138527 138738 8.85 + 2 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 138813 139032 3.82 + 0 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 139126 139281 0.68 + 2 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 139364 139531 2.81 + 2 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 139602 139739 0.68 + 2 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 139819 139941 2.96 + 2 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 140056 140276 4.70 + 2 SCAF14367_23 SCAF14367 geneid_v1.1 Terminal 140347 140571 7.97 + 0 SCAF14367_23 # Gene 24 (Reverse). 2 exons. 240 aa. Score = 5.945699 SCAF14367 geneid_v1.1 Terminal 141286 141993 8.12 - 0 SCAF14367_24 SCAF14367 geneid_v1.1 First 146084 146095 -2.17 - 0 SCAF14367_24 # Gene 25 (Forward). 8 exons. 500 aa. Score = 32.864351 SCAF14367 geneid_v1.1 First 146836 146898 -4.06 + 0 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 146960 147127 2.78 + 0 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 147324 147576 6.37 + 0 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 147767 148224 10.97 + 2 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 148530 148685 4.83 + 0 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 148892 149016 4.02 + 0 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 149096 149198 2.45 + 1 SCAF14367_25 SCAF14367 geneid_v1.1 Terminal 149393 149566 5.49 + 0 SCAF14367_25 # Gene 26 (Reverse). 8 exons. 407 aa. Score = 26.885440 SCAF14367 geneid_v1.1 Terminal 150338 150434 -0.50 - 1 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 150718 150824 0.28 - 0 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 150888 151261 14.46 - 2 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 151338 151525 4.06 - 1 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 151752 151828 0.40 - 0 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 151903 151997 0.52 - 2 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 152183 152353 5.62 - 2 SCAF14367_26 SCAF14367 geneid_v1.1 First 152425 152536 2.04 - 0 SCAF14367_26 # Gene 27 (Forward). 5 exons. 292 aa. Score = 10.672010 SCAF14367 geneid_v1.1 First 155472 155577 0.38 + 0 SCAF14367_27 SCAF14367 geneid_v1.1 Internal 155696 155849 0.54 + 2 SCAF14367_27 SCAF14367 geneid_v1.1 Internal 155937 156059 0.88 + 1 SCAF14367_27 SCAF14367 geneid_v1.1 Internal 158345 158371 0.13 + 1 SCAF14367_27 SCAF14367 geneid_v1.1 Terminal 158587 159052 8.74 + 1 SCAF14367_27 # Gene 28 (Reverse). 6 exons. 430 aa. Score = 22.151595 SCAF14367 geneid_v1.1 Terminal 160170 160322 1.37 - 0 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 160934 161122 2.99 - 0 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 161191 161370 4.10 - 0 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 161418 161614 5.79 - 2 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 162037 162426 7.30 - 2 SCAF14367_28 SCAF14367 geneid_v1.1 First 162699 162879 0.60 - 0 SCAF14367_28 # Gene 29 (Forward). 24 exons. 970 aa. Score = 62.646176 SCAF14367 geneid_v1.1 First 163512 163800 4.56 + 0 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 164897 165127 4.24 + 2 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 165389 165485 4.70 + 2 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 165618 165760 6.40 + 1 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 166001 166098 0.35 + 2 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 166209 166357 -0.97 + 0 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 166462 166625 1.80 + 1 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 168753 168784 0.75 + 2 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 168825 168959 0.29 + 0 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 170890 170947 0.77 + 0 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 171013 171084 3.83 + 2 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 171193 171324 5.04 + 2 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 171471 171535 -0.56 + 2 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 171612 171738 4.39 + 0 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 171825 171888 1.29 + 2 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 171970 172056 1.73 + 1 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 172128 172278 3.47 + 1 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 172433 172580 7.41 + 0 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 172985 173092 2.74 + 2 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 174099 174252 -0.18 + 2 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 174372 174471 3.25 + 1 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 174646 174775 5.46 + 0 SCAF14367_29 SCAF14367 geneid_v1.1 Internal 175196 175275 2.16 + 2 SCAF14367_29 SCAF14367 geneid_v1.1 Terminal 175551 175646 -0.27 + 0 SCAF14367_29 # Gene 30 (Reverse). 9 exons. 504 aa. Score = 60.823906 SCAF14367 geneid_v1.1 Terminal 177290 177556 12.24 - 0 SCAF14367_30 SCAF14367 geneid_v1.1 Internal 177881 178025 1.49 - 1 SCAF14367_30 SCAF14367 geneid_v1.1 Internal 178075 178209 3.17 - 1 SCAF14367_30 SCAF14367 geneid_v1.1 Internal 179196 179390 13.97 - 1 SCAF14367_30 SCAF14367 geneid_v1.1 Internal 179924 180157 6.97 - 1 SCAF14367_30 SCAF14367 geneid_v1.1 Internal 180331 180430 1.14 - 2 SCAF14367_30 SCAF14367 geneid_v1.1 Internal 180505 180736 11.21 - 0 SCAF14367_30 SCAF14367 geneid_v1.1 Internal 180953 180995 1.35 - 1 SCAF14367_30 SCAF14367 geneid_v1.1 First 182435 182595 9.28 - 0 SCAF14367_30 # Gene 31 (Forward). 1 exons. 36 aa. Score = 0.683949 SCAF14367 geneid_v1.1 Single 185540 185647 0.68 + 0 SCAF14367_31 # Gene 32 (Reverse). 5 exons. 297 aa. Score = 9.699179 SCAF14367 geneid_v1.1 Terminal 186413 186605 1.89 - 1 SCAF14367_32 SCAF14367 geneid_v1.1 Internal 186811 186890 5.66 - 0 SCAF14367_32 SCAF14367 geneid_v1.1 Internal 187437 187595 0.43 - 0 SCAF14367_32 SCAF14367 geneid_v1.1 Internal 190773 191196 1.86 - 1 SCAF14367_32 SCAF14367 geneid_v1.1 First 191773 191807 -0.15 - 0 SCAF14367_32 # Gene 33 (Forward). 7 exons. 596 aa. Score = 36.248812 SCAF14367 geneid_v1.1 First 193682 193860 10.75 + 0 SCAF14367_33 SCAF14367 geneid_v1.1 Internal 194063 194199 1.20 + 1 SCAF14367_33 SCAF14367 geneid_v1.1 Internal 196954 197335 14.61 + 2 SCAF14367_33 SCAF14367 geneid_v1.1 Internal 198054 198530 3.92 + 1 SCAF14367_33 SCAF14367 geneid_v1.1 Internal 198858 198978 -1.63 + 1 SCAF14367_33 SCAF14367 geneid_v1.1 Internal 202047 202342 8.83 + 0 SCAF14367_33 SCAF14367 geneid_v1.1 Terminal 202842 203037 -1.44 + 1 SCAF14367_33 # Gene 34 (Reverse). 3 exons. 116 aa. Score = 9.640206 SCAF14367 geneid_v1.1 Terminal 203923 204054 3.83 - 0 SCAF14367_34 SCAF14367 geneid_v1.1 Internal 205632 205799 7.40 - 0 SCAF14367_34 SCAF14367 geneid_v1.1 First 207037 207084 -1.60 - 0 SCAF14367_34 # Gene 35 (Forward). 25 exons. 1325 aa. Score = 113.661234 SCAF14367 geneid_v1.1 First 207956 208058 2.43 + 0 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 209570 209804 -0.07 + 2 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 217098 217353 12.40 + 1 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 217407 217531 0.78 + 0 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 217674 217813 0.75 + 1 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 217945 218092 2.78 + 2 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 218213 218370 6.48 + 1 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 218734 218867 4.29 + 2 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 221504 221731 3.28 + 0 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 225955 226119 10.24 + 0 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 226220 226355 7.24 + 0 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 228398 228552 8.07 + 2 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 229407 229536 5.00 + 0 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 229653 229798 4.09 + 2 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 229883 230394 10.01 + 0 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 231321 231447 3.66 + 1 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 231516 231652 4.70 + 0 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 231714 231789 -0.67 + 1 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 231892 231983 2.51 + 0 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 232061 232237 7.27 + 1 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 232317 232379 2.07 + 1 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 232452 232528 1.48 + 1 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 232650 232792 2.15 + 2 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 232870 232977 4.61 + 0 SCAF14367_35 SCAF14367 geneid_v1.1 Terminal 233215 233418 8.10 + 0 SCAF14367_35 # Gene 36 (Reverse). 7 exons. 319 aa. Score = 36.659185 SCAF14367 geneid_v1.1 Terminal 235142 235275 4.36 - 2 SCAF14367_36 SCAF14367 geneid_v1.1 Internal 235384 235665 14.40 - 2 SCAF14367_36 SCAF14367 geneid_v1.1 Internal 236050 236179 3.70 - 0 SCAF14367_36 SCAF14367 geneid_v1.1 Internal 236325 236471 8.65 - 0 SCAF14367_36 SCAF14367 geneid_v1.1 Internal 236641 236774 5.11 - 2 SCAF14367_36 SCAF14367 geneid_v1.1 Internal 237587 237699 1.36 - 1 SCAF14367_36 SCAF14367 geneid_v1.1 First 238440 238456 -0.93 - 0 SCAF14367_36 # Gene 37 (Forward). 2 exons. 921 aa. Score = 105.798113 SCAF14367 geneid_v1.1 First 239925 240138 1.89 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Terminal 241559 244107 103.90 + 2 SCAF14367_37 # Gene 38 (Forward). 8 exons. 370 aa. Score = 13.720825 SCAF14367 geneid_v1.1 First 246669 246759 -1.13 + 0 SCAF14367_38 SCAF14367 geneid_v1.1 Internal 246884 246977 -0.33 + 2 SCAF14367_38 SCAF14367 geneid_v1.1 Internal 247058 247451 9.16 + 1 SCAF14367_38 SCAF14367 geneid_v1.1 Internal 247524 247615 -4.06 + 0 SCAF14367_38 SCAF14367 geneid_v1.1 Internal 247876 248023 7.48 + 1 SCAF14367_38 SCAF14367 geneid_v1.1 Internal 248098 248158 0.27 + 0 SCAF14367_38 SCAF14367 geneid_v1.1 Internal 248241 248371 3.65 + 2 SCAF14367_38 SCAF14367 geneid_v1.1 Terminal 248464 248562 -1.32 + 0 SCAF14367_38 # Gene 39 (Reverse). 6 exons. 1161 aa. Score = 78.960982 SCAF14367 geneid_v1.1 Terminal 249464 249650 3.73 - 1 SCAF14367_39 SCAF14367 geneid_v1.1 Internal 249782 249884 -1.17 - 2 SCAF14367_39 SCAF14367 geneid_v1.1 Internal 250727 250852 0.87 - 2 SCAF14367_39 SCAF14367 geneid_v1.1 Internal 251434 252024 11.12 - 2 SCAF14367_39 SCAF14367 geneid_v1.1 Internal 252274 254643 64.60 - 2 SCAF14367_39 SCAF14367 geneid_v1.1 First 258619 258724 -0.18 - 0 SCAF14367_39 # Gene 40 (Forward). 6 exons. 323 aa. Score = 10.022258 SCAF14367 geneid_v1.1 First 259661 259894 -2.10 + 0 SCAF14367_40 SCAF14367 geneid_v1.1 Internal 260118 260273 1.89 + 0 SCAF14367_40 SCAF14367 geneid_v1.1 Internal 261567 261661 -0.08 + 0 SCAF14367_40 SCAF14367 geneid_v1.1 Internal 261775 261908 0.89 + 1 SCAF14367_40 SCAF14367 geneid_v1.1 Internal 262153 262233 1.96 + 2 SCAF14367_40 SCAF14367 geneid_v1.1 Terminal 266244 266512 7.46 + 2 SCAF14367_40 # Gene 41 (Forward). 6 exons. 391 aa. Score = 16.275506 SCAF14367 geneid_v1.1 First 268049 268268 2.51 + 0 SCAF14367_41 SCAF14367 geneid_v1.1 Internal 268840 268926 3.23 + 2 SCAF14367_41 SCAF14367 geneid_v1.1 Internal 269041 269176 -0.22 + 2 SCAF14367_41 SCAF14367 geneid_v1.1 Internal 269676 269942 3.96 + 1 SCAF14367_41 SCAF14367 geneid_v1.1 Internal 270845 270948 0.29 + 1 SCAF14367_41 SCAF14367 geneid_v1.1 Terminal 271032 271390 6.50 + 2 SCAF14367_41 # Gene 42 (Reverse). 8 exons. 449 aa. Score = 21.191504 SCAF14367 geneid_v1.1 Terminal 272056 272282 6.79 - 2 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 272684 272837 2.37 - 0 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 272923 273007 2.97 - 1 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 274048 274119 1.38 - 1 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 274274 274553 5.29 - 2 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 274622 274730 0.24 - 0 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 274804 274917 0.52 - 0 SCAF14367_42 SCAF14367 geneid_v1.1 First 274993 275298 1.63 - 0 SCAF14367_42 # Gene 43 (Forward). 6 exons. 293 aa. Score = 9.673016 SCAF14367 geneid_v1.1 First 276174 276278 0.44 + 0 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 276358 276419 -0.34 + 0 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 277055 277445 3.05 + 1 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 278000 278106 2.16 + 0 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 278984 279052 1.92 + 1 SCAF14367_43 SCAF14367 geneid_v1.1 Terminal 280186 280330 2.44 + 1 SCAF14367_43 # Gene 44 (Reverse). 5 exons. 343 aa. Score = 16.076450 SCAF14367 geneid_v1.1 Terminal 281269 281447 4.85 - 2 SCAF14367_44 SCAF14367 geneid_v1.1 Internal 281577 281709 1.23 - 0 SCAF14367_44 SCAF14367 geneid_v1.1 Internal 282015 282415 8.45 - 2 SCAF14367_44 SCAF14367 geneid_v1.1 Internal 282675 282878 1.10 - 2 SCAF14367_44 SCAF14367 geneid_v1.1 First 282956 283067 0.44 - 0 SCAF14367_44 # Gene 45 (Forward). 8 exons. 300 aa. Score = 40.438112 SCAF14367 geneid_v1.1 First 285362 285367 -0.75 + 0 SCAF14367_45 SCAF14367 geneid_v1.1 Internal 287909 287978 3.19 + 0 SCAF14367_45 SCAF14367 geneid_v1.1 Internal 288107 288222 5.73 + 2 SCAF14367_45 SCAF14367 geneid_v1.1 Internal 288305 288439 6.51 + 0 SCAF14367_45 SCAF14367 geneid_v1.1 Internal 288515 288717 6.17 + 0 SCAF14367_45 SCAF14367 geneid_v1.1 Internal 288986 289163 13.57 + 1 SCAF14367_45 SCAF14367 geneid_v1.1 Internal 289336 289424 5.70 + 0 SCAF14367_45 SCAF14367 geneid_v1.1 Terminal 289592 289694 0.32 + 1 SCAF14367_45 # Gene 46 (Reverse). 4 exons. 334 aa. Score = 35.354458 SCAF14367 geneid_v1.1 Terminal 290680 291360 26.58 - 0 SCAF14367_46 SCAF14367 geneid_v1.1 Internal 291449 291580 0.84 - 0 SCAF14367_46 SCAF14367 geneid_v1.1 Internal 292093 292227 5.80 - 0 SCAF14367_46 SCAF14367 geneid_v1.1 First 293517 293570 2.13 - 0 SCAF14367_46 # Gene 47 (Forward). 24 exons. 1221 aa. Score = 52.693133 SCAF14367 geneid_v1.1 First 294423 294487 -1.47 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 295426 295490 -2.09 + 1 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 295830 295943 1.73 + 2 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 296719 297070 2.10 + 2 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 297482 297761 5.67 + 1 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 297863 297930 0.98 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 298032 298202 2.83 + 1 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 298703 298801 1.91 + 1 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 303851 303947 3.62 + 1 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 304056 304177 1.49 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 304265 304544 7.74 + 1 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 304724 304811 -0.22 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 304961 305178 5.79 + 2 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 305603 305756 -0.32 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 305853 305965 2.80 + 2 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 307645 307787 1.88 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 308436 308599 2.37 + 1 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 308698 308911 7.12 + 2 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 308954 309083 -2.49 + 1 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 310380 310508 2.10 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 310585 310722 1.32 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 311628 311774 0.49 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 312007 312309 8.71 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Terminal 313977 313985 -1.37 + 0 SCAF14367_47 # Gene 48 (Forward). 14 exons. 722 aa. Score = 68.345568 SCAF14367 geneid_v1.1 First 316111 316291 4.77 + 0 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 316871 317025 9.50 + 2 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 318075 318236 4.53 + 0 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 318313 318462 4.62 + 0 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 318825 318917 3.49 + 0 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 319006 319225 5.12 + 0 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 319254 319342 3.44 + 2 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 319419 319634 12.61 + 0 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 319706 319839 3.66 + 0 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 319913 320003 1.74 + 1 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 320411 320530 0.46 + 0 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 320607 320759 6.35 + 0 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 321054 321131 4.27 + 0 SCAF14367_48 SCAF14367 geneid_v1.1 Terminal 321276 321599 3.79 + 0 SCAF14367_48 # Gene 49 (Forward). 2 exons. 140 aa. Score = 23.261977 SCAF14367 geneid_v1.1 First 322423 322528 4.51 + 0 SCAF14367_49 SCAF14367 geneid_v1.1 Terminal 324378 324691 18.75 + 2 SCAF14367_49 # Gene 50 (Forward). 8 exons. 463 aa. Score = 50.390311 SCAF14367 geneid_v1.1 First 325856 326215 13.76 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 326297 326433 5.58 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 327263 327601 15.96 + 1 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 327673 327718 1.11 + 1 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 328099 328242 3.25 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 328325 328408 2.93 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 328493 328661 8.54 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Terminal 328926 329035 -0.75 + 2 SCAF14367_50 # Gene 51 (Reverse). 2 exons. 234 aa. Score = 1.766014 SCAF14367 geneid_v1.1 Terminal 336277 336596 0.59 - 2 SCAF14367_51 SCAF14367 geneid_v1.1 First 337836 338217 1.18 - 0 SCAF14367_51 # Gene 52 (Reverse). 1 exons. 82 aa. Score = 4.340908 SCAF14367 geneid_v1.1 Single 343156 343401 4.34 - 0 SCAF14367_52 # Gene 53 (Reverse). 1 exons. 82 aa. Score = 4.341494 SCAF14367 geneid_v1.1 Single 347323 347568 4.34 - 0 SCAF14367_53 # Gene 54 (Forward). 2 exons. 73 aa. Score = 6.866810 SCAF14367 geneid_v1.1 First 349737 349829 4.77 + 0 SCAF14367_54 SCAF14367 geneid_v1.1 Terminal 350010 350135 2.10 + 0 SCAF14367_54 # Gene 55 (Reverse). 1 exons. 70 aa. Score = 1.016101 SCAF14367 geneid_v1.1 Single 351363 351572 1.02 - 0 SCAF14367_55 # Gene 56 (Reverse). 6 exons. 342 aa. Score = 8.745749 SCAF14367 geneid_v1.1 Terminal 354178 354291 0.44 - 0 SCAF14367_56 SCAF14367 geneid_v1.1 Internal 354571 354793 7.89 - 1 SCAF14367_56 SCAF14367 geneid_v1.1 Internal 354946 355047 0.14 - 1 SCAF14367_56 SCAF14367 geneid_v1.1 Internal 355536 355918 0.57 - 0 SCAF14367_56 SCAF14367 geneid_v1.1 Internal 362968 363142 0.53 - 1 SCAF14367_56 SCAF14367 geneid_v1.1 First 363247 363275 -0.82 - 0 SCAF14367_56 # Gene 57 (Reverse). 5 exons. 323 aa. Score = 8.113784 SCAF14367 geneid_v1.1 Terminal 364533 364770 3.94 - 1 SCAF14367_57 SCAF14367 geneid_v1.1 Internal 369353 369624 0.83 - 0 SCAF14367_57 SCAF14367 geneid_v1.1 Internal 369853 369959 1.66 - 2 SCAF14367_57 SCAF14367 geneid_v1.1 Internal 377094 377325 1.26 - 0 SCAF14367_57 SCAF14367 geneid_v1.1 First 378172 378291 0.43 - 0 SCAF14367_57 # Gene 58 (Forward). 4 exons. 325 aa. Score = 26.789396 SCAF14367 geneid_v1.1 First 379852 380005 9.82 + 0 SCAF14367_58 SCAF14367 geneid_v1.1 Internal 380108 380286 6.54 + 2 SCAF14367_58 SCAF14367 geneid_v1.1 Internal 380614 380920 4.63 + 0 SCAF14367_58 SCAF14367 geneid_v1.1 Terminal 381215 381549 5.80 + 2 SCAF14367_58 # Gene 59 (Reverse). 10 exons. 771 aa. Score = 39.137329 SCAF14367 geneid_v1.1 Terminal 387848 387976 1.50 - 0 SCAF14367_59 SCAF14367 geneid_v1.1 Internal 388060 388220 2.35 - 2 SCAF14367_59 SCAF14367 geneid_v1.1 Internal 388901 389075 4.18 - 0 SCAF14367_59 SCAF14367 geneid_v1.1 Internal 389304 389509 1.69 - 2 SCAF14367_59 SCAF14367 geneid_v1.1 Internal 390376 390859 -1.04 - 0 SCAF14367_59 SCAF14367 geneid_v1.1 Internal 391740 391931 5.73 - 0 SCAF14367_59 SCAF14367 geneid_v1.1 Internal 392653 392798 3.19 - 2 SCAF14367_59 SCAF14367 geneid_v1.1 Internal 393295 393434 2.43 - 1 SCAF14367_59 SCAF14367 geneid_v1.1 Internal 394820 394947 -0.29 - 0 SCAF14367_59 SCAF14367 geneid_v1.1 First 395028 395579 19.40 - 0 SCAF14367_59 # Gene 60 (Forward). 4 exons. 235 aa. Score = 3.137236 SCAF14367 geneid_v1.1 First 398237 398583 3.53 + 0 SCAF14367_60 SCAF14367 geneid_v1.1 Internal 399872 400027 0.18 + 1 SCAF14367_60 SCAF14367 geneid_v1.1 Internal 401736 401763 -1.05 + 1 SCAF14367_60 SCAF14367 geneid_v1.1 Terminal 402669 402842 0.47 + 0 SCAF14367_60 # Gene 61 (Forward). 2 exons. 450 aa. Score = 54.062539 SCAF14367 geneid_v1.1 First 403476 404291 36.68 + 0 SCAF14367_61 SCAF14367 geneid_v1.1 Terminal 404444 404977 17.38 + 0 SCAF14367_61 # Gene 62 (Reverse). 1 exons. 92 aa. Score = 12.449867 SCAF14367 geneid_v1.1 Single 406216 406491 12.45 - 0 SCAF14367_62 # Gene 63 (Forward). 2 exons. 154 aa. Score = 3.090237 SCAF14367 geneid_v1.1 First 408473 408895 4.53 + 0 SCAF14367_63 SCAF14367 geneid_v1.1 Terminal 409025 409063 -1.44 + 0 SCAF14367_63 # Gene 64 (Forward). 2 exons. 72 aa. Score = 3.289718 SCAF14367 geneid_v1.1 First 412979 413040 2.21 + 0 SCAF14367_64 SCAF14367 geneid_v1.1 Terminal 414391 414544 1.08 + 1 SCAF14367_64 # Gene 65 (Forward). 2 exons. 73 aa. Score = 0.905785 SCAF14367 geneid_v1.1 First 417653 417665 -0.71 + 0 SCAF14367_65 SCAF14367 geneid_v1.1 Terminal 420241 420446 1.62 + 2 SCAF14367_65 # Gene 66 (Forward). 1 exons. 26 aa. Score = 0.363535 SCAF14367 geneid_v1.1 Single 421911 421988 0.36 + 0 SCAF14367_66 # Gene 67 (Reverse). 12 exons. 519 aa. Score = 31.539101 SCAF14367 geneid_v1.1 Terminal 423427 423516 2.92 - 0 SCAF14367_67 SCAF14367 geneid_v1.1 Internal 424051 424197 10.58 - 0 SCAF14367_67 SCAF14367 geneid_v1.1 Internal 425768 425914 1.54 - 0 SCAF14367_67 SCAF14367 geneid_v1.1 Internal 428889 429060 -1.16 - 1 SCAF14367_67 SCAF14367 geneid_v1.1 Internal 430736 431049 2.19 - 0 SCAF14367_67 SCAF14367 geneid_v1.1 Internal 432852 433038 7.09 - 1 SCAF14367_67 SCAF14367 geneid_v1.1 Internal 433123 433218 3.14 - 1 SCAF14367_67 SCAF14367 geneid_v1.1 Internal 433287 433350 0.69 - 2 SCAF14367_67 SCAF14367 geneid_v1.1 Internal 436811 436905 -0.01 - 1 SCAF14367_67 SCAF14367 geneid_v1.1 Internal 439379 439471 2.38 - 1 SCAF14367_67 SCAF14367 geneid_v1.1 Internal 440697 440739 1.29 - 2 SCAF14367_67 SCAF14367 geneid_v1.1 First 444708 444816 0.90 - 0 SCAF14367_67
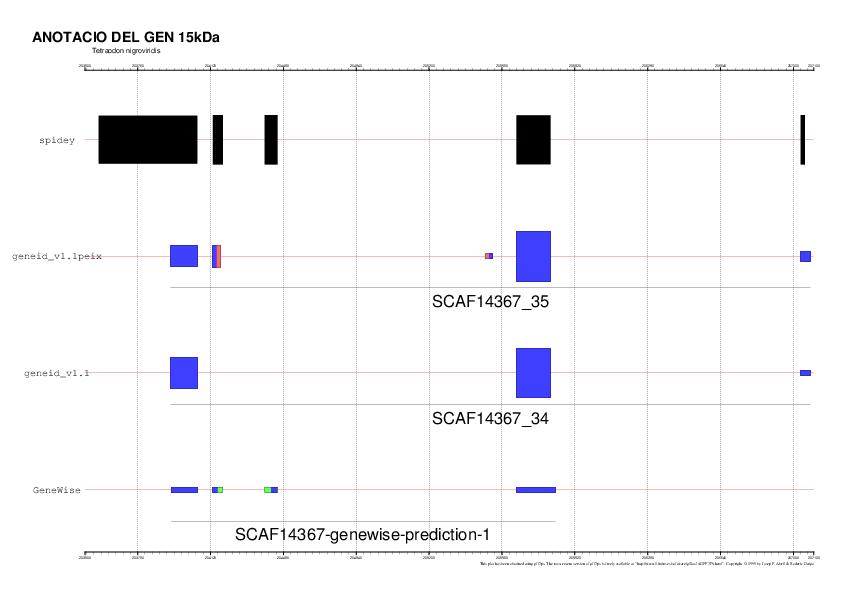
El gen que surt de color vermell -gen 34- és el que correspon amb el gen 15kDa
## gff-version 2 ## date Mon Mar 8 18:48:37 2004 ## source-version: geneid v 1.1 -- geneid@imim.es # Sequence SCAF14367 - Length = 444931 bps # Optimal Gene Structure. 72 genes. Score = 2039.635169 # Gene 1 (Reverse). 9 exons. 343 aa. Score = 37.668026 SCAF14367 geneid_v1.1 Internal 2912 3090 10.85 - 0 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 3172 3338 6.50 - 2 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 3513 3606 1.01 - 0 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 3972 4098 -0.56 - 1 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 5827 5999 8.96 - 0 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 6077 6243 8.60 - 2 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 6314 6393 0.77 - 1 SCAF14367_1 SCAF14367 geneid_v1.1 Internal 6563 6584 1.74 - 2 SCAF14367_1 SCAF14367 geneid_v1.1 First 8520 8538 -0.19 - 0 SCAF14367_1 # Gene 2 (Reverse). 5 exons. 214 aa. Score = 27.705650 SCAF14367 geneid_v1.1 Terminal 9582 9836 10.58 - 0 SCAF14367_2 SCAF14367 geneid_v1.1 Internal 10509 10675 11.97 - 2 SCAF14367_2 SCAF14367 geneid_v1.1 Internal 11010 11071 2.41 - 1 SCAF14367_2 SCAF14367 geneid_v1.1 Internal 11188 11292 1.38 - 1 SCAF14367_2 SCAF14367 geneid_v1.1 First 11496 11548 1.37 - 0 SCAF14367_2 # Gene 3 (Forward). 5 exons. 94 aa. Score = 2.044189 SCAF14367 geneid_v1.1 First 19696 19789 0.29 + 0 SCAF14367_3 SCAF14367 geneid_v1.1 Internal 21266 21385 2.50 + 2 SCAF14367_3 SCAF14367 geneid_v1.1 Internal 23252 23280 -0.53 + 2 SCAF14367_3 SCAF14367 geneid_v1.1 Internal 23406 23440 1.18 + 0 SCAF14367_3 SCAF14367 geneid_v1.1 Terminal 27584 27587 -1.39 + 1 SCAF14367_3 # Gene 4 (Forward). 10 exons. 752 aa. Score = 82.124930 SCAF14367 geneid_v1.1 First 30851 30952 1.31 + 0 SCAF14367_4 SCAF14367 geneid_v1.1 Internal 32166 32293 0.12 + 0 SCAF14367_4 SCAF14367 geneid_v1.1 Internal 35730 35906 7.11 + 1 SCAF14367_4 SCAF14367 geneid_v1.1 Internal 38840 39042 6.81 + 1 SCAF14367_4 SCAF14367 geneid_v1.1 Internal 41410 41633 6.48 + 2 SCAF14367_4 SCAF14367 geneid_v1.1 Internal 43215 43320 3.58 + 0 SCAF14367_4 SCAF14367 geneid_v1.1 Internal 43490 43726 11.74 + 2 SCAF14367_4 SCAF14367 geneid_v1.1 Internal 44585 44807 10.19 + 2 SCAF14367_4 SCAF14367 geneid_v1.1 Internal 45858 45918 2.61 + 1 SCAF14367_4 SCAF14367 geneid_v1.1 Terminal 46187 46981 32.17 + 0 SCAF14367_4 # Gene 5 (Forward). 12 exons. 513 aa. Score = 21.374634 SCAF14367 geneid_v1.1 First 48707 48784 0.97 + 0 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 49867 49929 0.19 + 0 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 52398 52484 0.75 + 0 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 52588 52634 0.58 + 0 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 52861 53002 2.86 + 1 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 53088 53273 4.98 + 0 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 54194 54390 4.64 + 0 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 54463 54576 5.84 + 1 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 55798 56144 0.83 + 1 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 56697 56766 0.15 + 2 SCAF14367_5 SCAF14367 geneid_v1.1 Internal 57157 57175 -0.25 + 1 SCAF14367_5 SCAF14367 geneid_v1.1 Terminal 61626 61814 -0.16 + 0 SCAF14367_5 # Gene 6 (Forward). 14 exons. 654 aa. Score = 93.005762 SCAF14367 geneid_v1.1 First 64853 64896 1.06 + 0 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 64977 65066 5.28 + 1 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 65139 65200 1.07 + 1 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 65279 65682 26.34 + 2 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 66477 66544 -1.12 + 0 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 70325 70460 5.85 + 1 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 70899 70970 2.58 + 0 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 72110 72202 6.77 + 0 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 72277 72335 1.50 + 0 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 72406 72514 10.16 + 1 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 72857 72887 0.12 + 0 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 73112 73233 5.42 + 2 SCAF14367_6 SCAF14367 geneid_v1.1 Internal 74613 74781 7.43 + 0 SCAF14367_6 SCAF14367 geneid_v1.1 Terminal 75195 75697 20.56 + 2 SCAF14367_6 # Gene 7 (Forward). 16 exons. 798 aa. Score = 77.703576 SCAF14367 geneid_v1.1 First 77616 77700 2.39 + 0 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 78738 78810 1.85 + 2 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 79205 79442 13.56 + 1 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 79551 79684 8.96 + 0 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 79999 80182 5.70 + 1 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 80274 80514 3.40 + 0 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 80587 80652 0.01 + 2 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 80844 80980 3.38 + 2 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 81065 81266 5.45 + 0 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 81382 81560 3.76 + 2 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 81690 81817 6.23 + 0 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 81906 82124 8.68 + 1 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 82198 82432 8.57 + 1 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 82546 82695 6.35 + 0 SCAF14367_7 SCAF14367 geneid_v1.1 Internal 85219 85275 1.78 + 0 SCAF14367_7 SCAF14367 geneid_v1.1 Terminal 85781 85846 -2.38 + 0 SCAF14367_7 # Gene 8 (Forward). 5 exons. 249 aa. Score = 36.799251 SCAF14367 geneid_v1.1 First 86227 86482 13.08 + 0 SCAF14367_8 SCAF14367 geneid_v1.1 Internal 87166 87346 7.58 + 2 SCAF14367_8 SCAF14367 geneid_v1.1 Internal 87663 87734 2.41 + 1 SCAF14367_8 SCAF14367 geneid_v1.1 Internal 88148 88325 13.47 + 1 SCAF14367_8 SCAF14367 geneid_v1.1 Terminal 89399 89458 0.26 + 0 SCAF14367_8 # Gene 9 (Forward). 6 exons. 173 aa. Score = 11.412235 SCAF14367 geneid_v1.1 First 91727 91911 5.73 + 0 SCAF14367_9 SCAF14367 geneid_v1.1 Internal 92527 92560 1.20 + 1 SCAF14367_9 SCAF14367 geneid_v1.1 Internal 92635 92721 3.05 + 0 SCAF14367_9 SCAF14367 geneid_v1.1 Internal 92792 92859 0.99 + 0 SCAF14367_9 SCAF14367 geneid_v1.1 Internal 92944 93015 1.38 + 1 SCAF14367_9 SCAF14367 geneid_v1.1 Terminal 93087 93159 -0.93 + 1 SCAF14367_9 # Gene 10 (Forward). 6 exons. 156 aa. Score = 20.760665 SCAF14367 geneid_v1.1 First 93293 93355 5.18 + 0 SCAF14367_10 SCAF14367 geneid_v1.1 Internal 93420 93512 4.71 + 0 SCAF14367_10 SCAF14367 geneid_v1.1 Internal 93587 93654 1.93 + 0 SCAF14367_10 SCAF14367 geneid_v1.1 Internal 93724 93802 5.63 + 1 SCAF14367_10 SCAF14367 geneid_v1.1 Internal 93863 93945 2.36 + 0 SCAF14367_10 SCAF14367 geneid_v1.1 Terminal 94015 94096 0.96 + 1 SCAF14367_10 # Gene 11 (Reverse). 12 exons. 420 aa. Score = 34.939060 SCAF14367 geneid_v1.1 Terminal 95811 96115 5.49 - 2 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 96198 96325 5.74 - 1 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 96403 96449 -1.33 - 0 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 96660 96754 3.76 - 2 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 96840 96921 2.57 - 0 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 97020 97171 4.19 - 2 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 97269 97341 2.62 - 0 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 97442 97506 1.80 - 2 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 97594 97723 3.15 - 0 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 98123 98271 3.84 - 2 SCAF14367_11 SCAF14367 geneid_v1.1 Internal 98497 98521 1.06 - 0 SCAF14367_11 SCAF14367 geneid_v1.1 First 99231 99239 2.04 - 0 SCAF14367_11 # Gene 12 (Reverse). 10 exons. 440 aa. Score = 26.437755 SCAF14367 geneid_v1.1 Terminal 100808 101011 0.92 - 0 SCAF14367_12 SCAF14367 geneid_v1.1 Internal 101105 101229 1.54 - 2 SCAF14367_12 SCAF14367 geneid_v1.1 Internal 101311 101436 1.16 - 2 SCAF14367_12 SCAF14367 geneid_v1.1 Internal 103846 103980 1.15 - 2 SCAF14367_12 SCAF14367 geneid_v1.1 Internal 104087 104151 2.41 - 1 SCAF14367_12 SCAF14367 geneid_v1.1 Internal 104225 104427 8.61 - 0 SCAF14367_12 SCAF14367 geneid_v1.1 Internal 104499 104631 2.40 - 1 SCAF14367_12 SCAF14367 geneid_v1.1 Internal 104706 104861 2.92 - 1 SCAF14367_12 SCAF14367 geneid_v1.1 Internal 104960 105074 3.66 - 2 SCAF14367_12 SCAF14367 geneid_v1.1 First 105368 105425 1.69 - 0 SCAF14367_12 # Gene 13 (Forward). 1 exons. 27 aa. Score = 0.626164 SCAF14367 geneid_v1.1 Single 105608 105688 0.63 + 0 SCAF14367_13 # Gene 14 (Reverse). 8 exons. 466 aa. Score = 38.005733 SCAF14367 geneid_v1.1 Terminal 105954 106615 17.06 - 2 SCAF14367_14 SCAF14367 geneid_v1.1 Internal 106841 106915 3.26 - 2 SCAF14367_14 SCAF14367 geneid_v1.1 Internal 106965 107050 1.17 - 1 SCAF14367_14 SCAF14367 geneid_v1.1 Internal 107121 107184 3.83 - 2 SCAF14367_14 SCAF14367 geneid_v1.1 Internal 107262 107286 -1.23 - 0 SCAF14367_14 SCAF14367 geneid_v1.1 Internal 107800 108111 13.90 - 0 SCAF14367_14 SCAF14367 geneid_v1.1 Internal 108322 108487 1.15 - 1 SCAF14367_14 SCAF14367 geneid_v1.1 First 108990 108997 -1.15 - 0 SCAF14367_14 # Gene 15 (Forward). 2 exons. 107 aa. Score = 2.475209 SCAF14367 geneid_v1.1 First 109167 109255 -0.63 + 0 SCAF14367_15 SCAF14367 geneid_v1.1 Terminal 109530 109761 3.10 + 1 SCAF14367_15 # Gene 16 (Forward). 16 exons. 763 aa. Score = 52.983792 SCAF14367 geneid_v1.1 First 118294 118553 0.41 + 0 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 122693 122841 0.39 + 1 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 125195 125256 -0.36 + 2 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 126929 127103 0.81 + 0 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 128275 128373 2.02 + 2 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 129972 130092 2.97 + 2 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 130178 130226 3.05 + 1 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 130370 130552 10.71 + 0 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 130633 130749 4.07 + 0 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 130836 130978 5.22 + 0 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 131083 131259 8.19 + 1 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 131328 131402 3.60 + 1 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 131491 131661 1.95 + 1 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 131745 132008 7.00 + 1 SCAF14367_16 SCAF14367 geneid_v1.1 Internal 135468 135592 -0.34 + 1 SCAF14367_16 SCAF14367 geneid_v1.1 Terminal 136084 136202 3.31 + 2 SCAF14367_16 # Gene 17 (Forward). 9 exons. 598 aa. Score = 63.070483 SCAF14367 geneid_v1.1 First 138129 138459 15.10 + 0 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 138527 138738 13.82 + 2 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 138813 139032 3.05 + 0 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 139126 139281 2.96 + 2 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 139364 139531 4.60 + 2 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 139602 139739 2.11 + 2 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 139819 139941 4.35 + 2 SCAF14367_17 SCAF14367 geneid_v1.1 Internal 140056 140276 9.21 + 2 SCAF14367_17 SCAF14367 geneid_v1.1 Terminal 140347 140571 7.87 + 0 SCAF14367_17 # Gene 18 (Reverse). 2 exons. 262 aa. Score = 6.252652 SCAF14367 geneid_v1.1 Terminal 141286 141993 6.14 - 0 SCAF14367_18 SCAF14367 geneid_v1.1 First 144851 144928 0.11 - 0 SCAF14367_18 # Gene 19 (Forward). 10 exons. 505 aa. Score = 44.594113 SCAF14367 geneid_v1.1 First 145735 145794 0.98 + 0 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 146960 147089 4.21 + 0 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 147182 147237 -0.92 + 2 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 147324 147500 7.84 + 0 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 147605 147680 -1.43 + 0 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 147767 148224 10.22 + 2 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 148530 148685 4.36 + 0 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 148892 149016 5.46 + 0 SCAF14367_19 SCAF14367 geneid_v1.1 Internal 149096 149198 5.44 + 1 SCAF14367_19 SCAF14367 geneid_v1.1 Terminal 149393 149566 8.43 + 0 SCAF14367_19 # Gene 20 (Reverse). 6 exons. 279 aa. Score = 25.169027 SCAF14367 geneid_v1.1 Terminal 150714 150824 -1.75 - 0 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 150888 151261 12.85 - 2 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 151338 151493 5.26 - 2 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 151603 151676 3.87 - 1 SCAF14367_20 SCAF14367 geneid_v1.1 Internal 151752 151828 3.20 - 0 SCAF14367_20 SCAF14367 geneid_v1.1 First 151903 151947 1.74 - 0 SCAF14367_20 # Gene 21 (Reverse). 2 exons. 136 aa. Score = 12.315583 SCAF14367 geneid_v1.1 Terminal 152058 152353 8.39 - 2 SCAF14367_21 SCAF14367 geneid_v1.1 First 152425 152536 3.93 - 0 SCAF14367_21 # Gene 22 (Reverse). 1 exons. 35 aa. Score = 1.415576 SCAF14367 geneid_v1.1 Single 152896 153000 1.42 - 0 SCAF14367_22 # Gene 23 (Forward). 5 exons. 199 aa. Score = 13.188678 SCAF14367 geneid_v1.1 First 155472 155577 2.33 + 0 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 155696 155849 1.66 + 2 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 155937 156059 6.02 + 1 SCAF14367_23 SCAF14367 geneid_v1.1 Internal 156213 156343 2.14 + 1 SCAF14367_23 SCAF14367 geneid_v1.1 Terminal 156700 156782 1.04 + 2 SCAF14367_23 # Gene 24 (Forward). 5 exons. 234 aa. Score = 10.231257 SCAF14367 geneid_v1.1 First 157410 157460 -0.15 + 0 SCAF14367_24 SCAF14367 geneid_v1.1 Internal 157719 157803 0.94 + 0 SCAF14367_24 SCAF14367 geneid_v1.1 Internal 157882 157945 -0.44 + 2 SCAF14367_24 SCAF14367 geneid_v1.1 Internal 158587 159040 12.75 + 1 SCAF14367_24 SCAF14367 geneid_v1.1 Terminal 159718 159765 -2.87 + 0 SCAF14367_24 # Gene 25 (Reverse). 8 exons. 351 aa. Score = 24.718044 SCAF14367 geneid_v1.1 Terminal 160170 160290 2.25 - 1 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 160540 160681 5.74 - 2 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 160772 160859 -2.32 - 0 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 160934 161122 2.92 - 0 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 161191 161331 3.65 - 0 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 161418 161614 6.07 - 2 SCAF14367_25 SCAF14367 geneid_v1.1 Internal 162037 162166 4.77 - 0 SCAF14367_25 SCAF14367 geneid_v1.1 First 162281 162325 1.63 - 0 SCAF14367_25 # Gene 26 (Forward). 8 exons. 273 aa. Score = 20.492954 SCAF14367 geneid_v1.1 First 163512 163616 2.56 + 0 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 164731 164760 0.29 + 0 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 164968 165127 7.04 + 0 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 165389 165485 1.80 + 2 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 165618 165760 2.64 + 1 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 166001 166098 2.19 + 2 SCAF14367_26 SCAF14367 geneid_v1.1 Internal 166209 166257 0.57 + 0 SCAF14367_26 SCAF14367 geneid_v1.1 Terminal 166462 166598 3.41 + 2 SCAF14367_26 # Gene 27 (Reverse). 1 exons. 48 aa. Score = 0.427095 SCAF14367 geneid_v1.1 Single 168828 168971 0.43 - 0 SCAF14367_27 # Gene 28 (Forward). 12 exons. 372 aa. Score = 44.544389 SCAF14367 geneid_v1.1 First 170244 170300 1.49 + 0 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 170890 170947 3.69 + 0 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 171013 171084 2.37 + 2 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 171193 171324 5.09 + 2 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 171471 171535 1.02 + 2 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 171612 171738 5.64 + 0 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 171825 171888 0.85 + 2 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 171970 172056 4.08 + 1 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 172128 172278 7.35 + 1 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 172433 172580 10.42 + 0 SCAF14367_28 SCAF14367 geneid_v1.1 Internal 172985 173092 3.35 + 2 SCAF14367_28 SCAF14367 geneid_v1.1 Terminal 173302 173348 -0.81 + 2 SCAF14367_28 # Gene 29 (Reverse). 2 exons. 45 aa. Score = 0.672402 SCAF14367 geneid_v1.1 Terminal 173466 173581 0.23 - 2 SCAF14367_29 SCAF14367 geneid_v1.1 First 173758 173776 0.44 - 0 SCAF14367_29 # Gene 30 (Forward). 5 exons. 188 aa. Score = 21.907297 SCAF14367 geneid_v1.1 First 174083 174252 5.39 + 0 SCAF14367_30 SCAF14367 geneid_v1.1 Internal 174372 174471 3.92 + 1 SCAF14367_30 SCAF14367 geneid_v1.1 Internal 174646 174775 6.22 + 0 SCAF14367_30 SCAF14367 geneid_v1.1 Internal 175196 175275 2.75 + 2 SCAF14367_30 SCAF14367 geneid_v1.1 Terminal 175563 175646 3.63 + 0 SCAF14367_30 # Gene 31 (Reverse). 13 exons. 568 aa. Score = 87.425268 SCAF14367 geneid_v1.1 Terminal 177290 177556 12.18 - 0 SCAF14367_31 SCAF14367 geneid_v1.1 Internal 177881 177980 6.33 - 1 SCAF14367_31 SCAF14367 geneid_v1.1 Internal 178075 178209 3.44 - 1 SCAF14367_31 SCAF14367 geneid_v1.1 Internal 179196 179390 16.17 - 1 SCAF14367_31 SCAF14367 geneid_v1.1 Internal 179924 180157 10.30 - 1 SCAF14367_31 SCAF14367 geneid_v1.1 Internal 180331 180430 1.26 - 2 SCAF14367_31 SCAF14367 geneid_v1.1 Internal 180505 180736 14.82 - 0 SCAF14367_31 SCAF14367 geneid_v1.1 Internal 180953 180995 1.79 - 1 SCAF14367_31 SCAF14367 geneid_v1.1 Internal 182435 182639 16.20 - 2 SCAF14367_31 SCAF14367 geneid_v1.1 Internal 185982 186040 -0.74 - 1 SCAF14367_31 SCAF14367 geneid_v1.1 Internal 186811 186890 5.82 - 0 SCAF14367_31 SCAF14367 geneid_v1.1 Internal 190334 190366 1.18 - 0 SCAF14367_31 SCAF14367 geneid_v1.1 First 190486 190506 -1.33 - 0 SCAF14367_31 # Gene 32 (Forward). 3 exons. 234 aa. Score = 27.141463 SCAF14367 geneid_v1.1 First 193682 193860 11.66 + 0 SCAF14367_32 SCAF14367 geneid_v1.1 Internal 194063 194199 1.55 + 1 SCAF14367_32 SCAF14367 geneid_v1.1 Terminal 196954 197339 13.93 + 2 SCAF14367_32 # Gene 33 (Reverse). 2 exons. 211 aa. Score = 20.496858 SCAF14367 geneid_v1.1 Terminal 197836 197928 -3.68 - 0 SCAF14367_33 SCAF14367 geneid_v1.1 First 198010 198549 24.17 - 0 SCAF14367_33 # Gene 34 (Forward). 2 exons. 107 aa. Score = 8.404087 SCAF14367 geneid_v1.1 First 202116 202342 9.88 + 0 SCAF14367_34 SCAF14367 geneid_v1.1 Terminal 202740 202833 -1.48 + 1 SCAF14367_34 # Gene 35 (Reverse). 5 exons. 142 aa. Score = 14.281788 SCAF14367 geneid_v1.1 Terminal 203923 204054 2.69 - 0 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 204132 204171 3.01 - 1 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 205478 205515 -2.56 - 0 SCAF14367_35 SCAF14367 geneid_v1.1 Internal 205632 205799 12.11 - 0 SCAF14367_35 SCAF14367 geneid_v1.1 First 207037 207084 -0.97 - 0 SCAF14367_35 # Gene 36 (Forward). 8 exons. 432 aa. Score = 31.123095 SCAF14367 geneid_v1.1 First 207956 208058 5.87 + 0 SCAF14367_36 SCAF14367 geneid_v1.1 Internal 209570 209804 1.79 + 2 SCAF14367_36 SCAF14367 geneid_v1.1 Internal 212201 212309 -0.79 + 1 SCAF14367_36 SCAF14367 geneid_v1.1 Internal 216507 216631 -1.03 + 0 SCAF14367_36 SCAF14367 geneid_v1.1 Internal 217098 217353 10.75 + 1 SCAF14367_36 SCAF14367 geneid_v1.1 Internal 217446 217531 -1.00 + 0 SCAF14367_36 SCAF14367 geneid_v1.1 Internal 218213 218370 7.85 + 1 SCAF14367_36 SCAF14367 geneid_v1.1 Terminal 218734 218957 7.68 + 2 SCAF14367_36 # Gene 37 (Forward). 24 exons. 958 aa. Score = 99.358519 SCAF14367 geneid_v1.1 First 221711 221731 1.94 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 225955 226119 14.09 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 226220 226355 10.03 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 228398 228552 9.38 + 2 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 229407 229536 3.45 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 229653 229798 4.39 + 2 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 229883 230096 8.20 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 230209 230394 0.71 + 2 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 230486 230595 3.70 + 2 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 230687 230830 3.63 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 230901 230976 0.79 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 231090 231148 2.56 + 2 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 231197 231246 -1.04 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 231321 231447 4.95 + 1 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 231519 231652 7.28 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 231714 231789 -1.23 + 1 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 231892 231983 5.05 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 232061 232237 7.56 + 1 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 232317 232379 1.88 + 1 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 232452 232528 -0.45 + 1 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 232650 232792 5.83 + 2 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 232870 232977 4.77 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Internal 233055 233135 0.33 + 0 SCAF14367_37 SCAF14367 geneid_v1.1 Terminal 233215 233418 1.56 + 0 SCAF14367_37 # Gene 38 (Reverse). 7 exons. 319 aa. Score = 41.006995 SCAF14367 geneid_v1.1 Terminal 235142 235275 -0.08 - 2 SCAF14367_38 SCAF14367 geneid_v1.1 Internal 235384 235665 18.07 - 2 SCAF14367_38 SCAF14367 geneid_v1.1 Internal 236050 236179 3.95 - 0 SCAF14367_38 SCAF14367 geneid_v1.1 Internal 236325 236471 6.30 - 0 SCAF14367_38 SCAF14367 geneid_v1.1 Internal 236641 236774 5.90 - 2 SCAF14367_38 SCAF14367 geneid_v1.1 Internal 237587 237699 5.62 - 1 SCAF14367_38 SCAF14367 geneid_v1.1 First 238440 238456 1.25 - 0 SCAF14367_38 # Gene 39 (Forward). 3 exons. 870 aa. Score = 113.506754 SCAF14367 geneid_v1.1 First 239097 239119 -2.65 + 0 SCAF14367_39 SCAF14367 geneid_v1.1 Internal 239487 239524 2.15 + 1 SCAF14367_39 SCAF14367 geneid_v1.1 Terminal 241559 244107 114.01 + 2 SCAF14367_39 # Gene 40 (Forward). 8 exons. 351 aa. Score = 13.665152 SCAF14367 geneid_v1.1 First 246669 246759 0.59 + 0 SCAF14367_40 SCAF14367 geneid_v1.1 Internal 246884 246977 -1.21 + 2 SCAF14367_40 SCAF14367 geneid_v1.1 Internal 247058 247419 4.42 + 1 SCAF14367_40 SCAF14367 geneid_v1.1 Internal 247549 247615 -1.21 + 2 SCAF14367_40 SCAF14367 geneid_v1.1 Internal 247876 248023 6.44 + 1 SCAF14367_40 SCAF14367 geneid_v1.1 Internal 248098 248158 0.71 + 0 SCAF14367_40 SCAF14367 geneid_v1.1 Internal 248241 248371 5.35 + 2 SCAF14367_40 SCAF14367 geneid_v1.1 Terminal 248464 248562 -1.42 + 0 SCAF14367_40 # Gene 41 (Reverse). 7 exons. 756 aa. Score = 20.087334 SCAF14367 geneid_v1.1 Terminal 249464 249650 1.26 - 1 SCAF14367_41 SCAF14367 geneid_v1.1 Internal 249782 249884 0.11 - 2 SCAF14367_41 SCAF14367 geneid_v1.1 Internal 250670 250852 2.81 - 2 SCAF14367_41 SCAF14367 geneid_v1.1 Internal 251434 251652 0.96 - 2 SCAF14367_41 SCAF14367 geneid_v1.1 Internal 252274 252945 4.69 - 2 SCAF14367_41 SCAF14367 geneid_v1.1 Internal 253846 254674 11.82 - 0 SCAF14367_41 SCAF14367 geneid_v1.1 First 257675 257749 -1.55 - 0 SCAF14367_41 # Gene 42 (Forward). 17 exons. 593 aa. Score = 22.333705 SCAF14367 geneid_v1.1 First 258630 258750 1.29 + 0 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 259465 259513 0.46 + 2 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 261268 261403 0.59 + 1 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 261567 261661 1.45 + 0 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 261775 261908 3.24 + 1 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 262153 262233 3.42 + 2 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 262344 262435 1.72 + 2 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 262723 262757 -0.90 + 0 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 267468 267495 0.69 + 1 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 268540 268630 0.15 + 0 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 268840 268926 4.03 + 2 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 269486 269600 -0.58 + 2 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 269676 269739 2.58 + 1 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 270142 270249 2.43 + 0 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 270422 270455 -2.24 + 0 SCAF14367_42 SCAF14367 geneid_v1.1 Internal 270799 270948 2.17 + 2 SCAF14367_42 SCAF14367 geneid_v1.1 Terminal 271032 271390 1.84 + 2 SCAF14367_42 # Gene 43 (Reverse). 13 exons. 511 aa. Score = 25.920738 SCAF14367 geneid_v1.1 Terminal 272056 272145 1.77 - 0 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 272212 272282 4.07 - 2 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 272360 272495 3.74 - 0 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 272573 272598 -2.13 - 2 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 272684 272837 4.24 - 0 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 272923 273036 4.11 - 0 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 273380 273515 1.14 - 1 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 274048 274197 0.17 - 1 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 274274 274553 5.47 - 2 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 274622 274730 3.64 - 0 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 274804 274917 1.20 - 0 SCAF14367_43 SCAF14367 geneid_v1.1 Internal 274993 275118 0.57 - 0 SCAF14367_43 SCAF14367 geneid_v1.1 First 275272 275298 -2.07 - 0 SCAF14367_43 # Gene 44 (Forward). 6 exons. 200 aa. Score = 10.711838 SCAF14367 geneid_v1.1 First 276174 276278 0.89 + 0 SCAF14367_44 SCAF14367 geneid_v1.1 Internal 276358 276424 2.47 + 0 SCAF14367_44 SCAF14367 geneid_v1.1 Internal 277744 277850 1.68 + 2 SCAF14367_44 SCAF14367 geneid_v1.1 Internal 278000 278106 2.92 + 0 SCAF14367_44 SCAF14367 geneid_v1.1 Internal 278984 279052 1.31 + 1 SCAF14367_44 SCAF14367 geneid_v1.1 Terminal 280186 280330 1.45 + 1 SCAF14367_44 # Gene 45 (Reverse). 5 exons. 330 aa. Score = 19.175379 SCAF14367 geneid_v1.1 Terminal 281269 281447 6.16 - 2 SCAF14367_45 SCAF14367 geneid_v1.1 Internal 281542 281602 0.84 - 0 SCAF14367_45 SCAF14367 geneid_v1.1 Internal 282015 282415 4.87 - 2 SCAF14367_45 SCAF14367 geneid_v1.1 Internal 282675 282878 2.26 - 2 SCAF14367_45 SCAF14367 geneid_v1.1 First 282923 283067 5.05 - 0 SCAF14367_45 # Gene 46 (Reverse). 4 exons. 118 aa. Score = 0.780519 SCAF14367 geneid_v1.1 Terminal 284568 284714 -1.80 - 0 SCAF14367_46 SCAF14367 geneid_v1.1 Internal 285729 285845 0.62 - 0 SCAF14367_46 SCAF14367 geneid_v1.1 Internal 286384 286414 1.51 - 1 SCAF14367_46 SCAF14367 geneid_v1.1 First 286757 286815 0.45 - 0 SCAF14367_46 # Gene 47 (Forward). 8 exons. 299 aa. Score = 47.174630 SCAF14367 geneid_v1.1 First 287575 287577 -0.54 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 287909 287978 4.06 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 288107 288222 5.27 + 2 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 288305 288439 6.94 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 288515 288717 11.83 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 288986 289163 12.75 + 1 SCAF14367_47 SCAF14367 geneid_v1.1 Internal 289336 289424 7.49 + 0 SCAF14367_47 SCAF14367 geneid_v1.1 Terminal 289592 289694 -0.62 + 1 SCAF14367_47 # Gene 48 (Reverse). 5 exons. 510 aa. Score = 52.848395 SCAF14367 geneid_v1.1 Terminal 290680 291360 36.48 - 0 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 291449 291711 4.80 - 2 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 292053 292227 5.86 - 0 SCAF14367_48 SCAF14367 geneid_v1.1 Internal 292270 292626 0.07 - 0 SCAF14367_48 SCAF14367 geneid_v1.1 First 293517 293570 5.64 - 0 SCAF14367_48 # Gene 49 (Forward). 9 exons. 321 aa. Score = 13.861176 SCAF14367 geneid_v1.1 First 296246 296252 -1.93 + 0 SCAF14367_49 SCAF14367 geneid_v1.1 Internal 296719 296819 3.47 + 2 SCAF14367_49 SCAF14367 geneid_v1.1 Internal 296904 297070 1.71 + 0 SCAF14367_49 SCAF14367 geneid_v1.1 Internal 297181 297210 0.89 + 1 SCAF14367_49 SCAF14367 geneid_v1.1 Internal 297638 297761 4.76 + 1 SCAF14367_49 SCAF14367 geneid_v1.1 Internal 297863 297930 -0.19 + 0 SCAF14367_49 SCAF14367 geneid_v1.1 Internal 298032 298202 0.61 + 1 SCAF14367_49 SCAF14367 geneid_v1.1 Internal 298703 298801 0.45 + 1 SCAF14367_49 SCAF14367 geneid_v1.1 Terminal 299138 299333 4.09 + 1 SCAF14367_49 # Gene 50 (Forward). 14 exons. 611 aa. Score = 35.551246 SCAF14367 geneid_v1.1 First 302763 302837 1.86 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 303460 303617 -1.07 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 303851 303947 2.12 + 1 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 304056 304129 -0.10 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 304265 304544 9.85 + 1 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 304724 304811 -1.45 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 304961 305178 6.97 + 2 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 305603 305690 -0.62 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 305853 305965 3.54 + 2 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 306057 306134 0.93 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 306415 306453 0.86 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 307645 307787 6.18 + 0 SCAF14367_50 SCAF14367 geneid_v1.1 Internal 308436 308599 3.82 + 1 SCAF14367_50 SCAF14367 geneid_v1.1 Terminal 308698 308915 2.67 + 2 SCAF14367_50 # Gene 51 (Forward). 7 exons. 388 aa. Score = 29.665599 SCAF14367 geneid_v1.1 First 309716 309769 0.60 + 0 SCAF14367_51 SCAF14367 geneid_v1.1 Internal 309908 310045 7.48 + 0 SCAF14367_51 SCAF14367 geneid_v1.1 Internal 310380 310508 0.88 + 0 SCAF14367_51 SCAF14367 geneid_v1.1 Internal 310585 310722 8.33 + 0 SCAF14367_51 SCAF14367 geneid_v1.1 Internal 311133 311251 3.23 + 0 SCAF14367_51 SCAF14367 geneid_v1.1 Internal 311826 312309 9.51 + 1 SCAF14367_51 SCAF14367 geneid_v1.1 Terminal 315537 315638 -0.37 + 0 SCAF14367_51 # Gene 52 (Forward). 14 exons. 710 aa. Score = 95.167183 SCAF14367 geneid_v1.1 First 316111 316291 8.02 + 0 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 316871 317025 9.82 + 2 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 318075 318236 9.19 + 0 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 318313 318462 4.92 + 0 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 318825 318917 3.62 + 0 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 319006 319138 5.91 + 0 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 319254 319342 1.30 + 2 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 319419 319634 14.45 + 0 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 319706 319839 6.99 + 0 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 319913 320003 3.26 + 1 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 320360 320530 4.68 + 0 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 320607 320759 8.36 + 0 SCAF14367_52 SCAF14367 geneid_v1.1 Internal 321054 321131 6.88 + 0 SCAF14367_52 SCAF14367 geneid_v1.1 Terminal 321276 321599 7.78 + 0 SCAF14367_52 # Gene 53 (Forward). 3 exons. 164 aa. Score = 22.887200 SCAF14367 geneid_v1.1 First 322423 322528 4.16 + 0 SCAF14367_53 SCAF14367 geneid_v1.1 Internal 324378 324650 21.48 + 2 SCAF14367_53 SCAF14367 geneid_v1.1 Terminal 324919 325031 -2.76 + 2 SCAF14367_53 # Gene 54 (Reverse). 1 exons. 100 aa. Score = 3.000838 SCAF14367 geneid_v1.1 Single 325135 325434 3.00 - 0 SCAF14367_54 # Gene 55 (Forward). 8 exons. 465 aa. Score = 57.627286 SCAF14367 geneid_v1.1 First 325856 326215 16.94 + 0 SCAF14367_55 SCAF14367 geneid_v1.1 Internal 326297 326433 3.43 + 0 SCAF14367_55 SCAF14367 geneid_v1.1 Internal 327263 327601 18.63 + 1 SCAF14367_55 SCAF14367 geneid_v1.1 Internal 327673 327718 1.53 + 1 SCAF14367_55 SCAF14367 geneid_v1.1 Internal 328099 328242 5.33 + 0 SCAF14367_55 SCAF14367 geneid_v1.1 Internal 328325 328408 2.53 + 0 SCAF14367_55 SCAF14367 geneid_v1.1 Internal 328493 328661 9.12 + 0 SCAF14367_55 SCAF14367 geneid_v1.1 Terminal 329654 329769 0.12 + 2 SCAF14367_55 # Gene 56 (Forward). 3 exons. 94 aa. Score = 2.194316 SCAF14367 geneid_v1.1 First 329878 329926 0.90 + 0 SCAF14367_56 SCAF14367 geneid_v1.1 Internal 331302 331372 1.45 + 2 SCAF14367_56 SCAF14367 geneid_v1.1 Terminal 336115 336276 -0.16 + 0 SCAF14367_56 # Gene 57 (Reverse). 2 exons. 101 aa. Score = 2.739026 SCAF14367 geneid_v1.1 Terminal 341272 341355 -2.20 - 0 SCAF14367_57 SCAF14367 geneid_v1.1 First 343295 343513 4.94 - 0 SCAF14367_57 # Gene 58 (Reverse). 4 exons. 164 aa. Score = 3.611214 SCAF14367 geneid_v1.1 Terminal 344561 344649 -0.63 - 2 SCAF14367_58 SCAF14367 geneid_v1.1 Internal 346624 346694 1.41 - 1 SCAF14367_58 SCAF14367 geneid_v1.1 Internal 347113 347225 -1.92 - 0 SCAF14367_58 SCAF14367 geneid_v1.1 First 347462 347680 4.75 - 0 SCAF14367_58 # Gene 59 (Forward). 3 exons. 102 aa. Score = 8.675940 SCAF14367 geneid_v1.1 First 348011 348081 -0.41 + 0 SCAF14367_59 SCAF14367 geneid_v1.1 Internal 349721 349829 8.50 + 1 SCAF14367_59 SCAF14367 geneid_v1.1 Terminal 350010 350135 0.59 + 0 SCAF14367_59 # Gene 60 (Reverse). 1 exons. 50 aa. Score = 1.127479 SCAF14367 geneid_v1.1 Single 350355 350504 1.13 - 0 SCAF14367_60 # Gene 61 (Forward). 4 exons. 108 aa. Score = 0.457019 SCAF14367 geneid_v1.1 First 351339 351361 -0.94 + 0 SCAF14367_61 SCAF14367 geneid_v1.1 Internal 351649 351819 0.03 + 1 SCAF14367_61 SCAF14367 geneid_v1.1 Internal 353146 353218 1.49 + 1 SCAF14367_61 SCAF14367 geneid_v1.1 Terminal 353355 353411 -0.12 + 0 SCAF14367_61 # Gene 62 (Reverse). 4 exons. 160 aa. Score = 9.324297 SCAF14367 geneid_v1.1 Terminal 354178 354291 -0.73 - 0 SCAF14367_62 SCAF14367 geneid_v1.1 Internal 354571 354793 8.84 - 1 SCAF14367_62 SCAF14367 geneid_v1.1 Internal 354946 355047 0.94 - 1 SCAF14367_62 SCAF14367 geneid_v1.1 First 355536 355576 0.27 - 0 SCAF14367_62 # Gene 63 (Forward). 6 exons. 262 aa. Score = 2.070164 SCAF14367 geneid_v1.1 First 357704 357798 1.23 + 0 SCAF14367_63 SCAF14367 geneid_v1.1 Internal 357985 358010 0.23 + 1 SCAF14367_63 SCAF14367 geneid_v1.1 Internal 361488 361669 -0.98 + 2 SCAF14367_63 SCAF14367 geneid_v1.1 Internal 365035 365234 1.68 + 0 SCAF14367_63 SCAF14367 geneid_v1.1 Internal 367964 368076 0.60 + 1 SCAF14367_63 SCAF14367 geneid_v1.1 Terminal 369273 369442 -0.68 + 2 SCAF14367_63 # Gene 64 (Forward). 3 exons. 64 aa. Score = 4.809671 SCAF14367 geneid_v1.1 First 369603 369631 1.49 + 0 SCAF14367_64 SCAF14367 geneid_v1.1 Internal 374551 374615 4.33 + 1 SCAF14367_64 SCAF14367 geneid_v1.1 Terminal 377232 377329 -1.00 + 2 SCAF14367_64 # Gene 65 (Forward). 4 exons. 353 aa. Score = 44.742520 SCAF14367 geneid_v1.1 First 379852 380005 8.06 + 0 SCAF14367_65 SCAF14367 geneid_v1.1 Internal 380108 380286 11.77 + 2 SCAF14367_65 SCAF14367 geneid_v1.1 Internal 380614 381004 17.40 + 0 SCAF14367_65 SCAF14367 geneid_v1.1 Terminal 381215 381549 7.52 + 2 SCAF14367_65 # Gene 66 (Reverse). 6 exons. 263 aa. Score = 9.281614 SCAF14367 geneid_v1.1 Terminal 384376 384432 -0.73 - 0 SCAF14367_66 SCAF14367 geneid_v1.1 Internal 387637 387769 3.76 - 1 SCAF14367_66 SCAF14367 geneid_v1.1 Internal 387852 387976 1.98 - 0 SCAF14367_66 SCAF14367 geneid_v1.1 Internal 388060 388220 3.17 - 2 SCAF14367_66 SCAF14367 geneid_v1.1 Internal 388901 389120 3.06 - 0 SCAF14367_66 SCAF14367 geneid_v1.1 First 389304 389396 -1.96 - 0 SCAF14367_66 # Gene 67 (Forward). 2 exons. 74 aa. Score = 3.437872 SCAF14367 geneid_v1.1 First 389677 389724 2.48 + 0 SCAF14367_67 SCAF14367 geneid_v1.1 Terminal 390401 390574 0.96 + 0 SCAF14367_67 # Gene 68 (Reverse). 11 exons. 533 aa. Score = 56.970984 SCAF14367 geneid_v1.1 Terminal 391624 391668 -0.69 - 0 SCAF14367_68 SCAF14367 geneid_v1.1 Internal 391740 391931 9.36 - 0 SCAF14367_68 SCAF14367 geneid_v1.1 Internal 392653 392798 5.32 - 2 SCAF14367_68 SCAF14367 geneid_v1.1 Internal 392901 392988 0.12 - 0 SCAF14367_68 SCAF14367 geneid_v1.1 Internal 393055 393161 5.12 - 2 SCAF14367_68 SCAF14367 geneid_v1.1 Internal 393295 393434 4.04 - 1 SCAF14367_68 SCAF14367 geneid_v1.1 Internal 393574 393680 2.98 - 0 SCAF14367_68 SCAF14367 geneid_v1.1 Internal 393788 393862 2.35 - 0 SCAF14367_68 SCAF14367 geneid_v1.1 Internal 394039 394143 3.20 - 0 SCAF14367_68 SCAF14367 geneid_v1.1 Internal 394906 394947 0.23 - 0 SCAF14367_68 SCAF14367 geneid_v1.1 First 395028 395579 24.94 - 0 SCAF14367_68 # Gene 69 (Reverse). 2 exons. 28 aa. Score = 0.473088 SCAF14367 geneid_v1.1 Terminal 395758 395815 -0.63 - 1 SCAF14367_69 SCAF14367 geneid_v1.1 First 396531 396556 1.10 - 0 SCAF14367_69 # Gene 70 (Forward). 1 exons. 59 aa. Score = 0.151560 SCAF14367 geneid_v1.1 Single 401236 401412 0.15 + 0 SCAF14367_70 # Gene 71 (Forward). 2 exons. 450 aa. Score = 47.043029 SCAF14367 geneid_v1.1 First 403476 404291 35.51 + 0 SCAF14367_71 SCAF14367 geneid_v1.1 Terminal 404444 404977 11.53 + 0 SCAF14367_71 # Gene 72 (Reverse). 24 exons. 1041 aa. Score = 74.256149 SCAF14367 geneid_v1.1 Terminal 406216 406583 18.50 - 2 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 406917 407127 7.26 - 0 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 408312 408382 0.98 - 2 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 411371 411391 -1.68 - 2 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 415304 415384 1.52 - 2 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 416108 416219 0.44 - 0 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 416416 416535 1.18 - 0 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 416656 416710 1.34 - 1 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 418361 418615 3.57 - 1 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 419008 419237 1.06 - 0 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 419571 419660 0.21 - 0 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 420224 420430 2.37 - 0 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 421238 421489 1.64 - 0 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 423469 423516 2.56 - 0 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 424051 424293 11.94 - 0 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 425768 425863 4.33 - 0 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 429778 429798 -1.22 - 0 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 432852 433038 4.95 - 1 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 433123 433218 2.31 - 1 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 433287 433350 3.15 - 2 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 436536 436573 -0.53 - 1 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 439379 439471 5.24 - 1 SCAF14367_72 SCAF14367 geneid_v1.1 Internal 440697 440739 0.72 - 2 SCAF14367_72 SCAF14367 geneid_v1.1 First 444708 444828 2.43 - 0 SCAF14367_72
El gen que surt de color vermell -gen 35- és el que correspon amb el gen 15kDa

--SPIDEY version 1.40--
Genomic: lcl|SCAF14367 No definition line found, 444931 bp
mRNA: lcl|FD0ABA26AC03.contig No definition line found, 1123 bp
No alignment found.
--SPIDEY version 1.40--
Genomic: lcl|SCAF14367 No definition line found, 444931 bp
mRNA: lcl|FD0ADA46CC04.contig No definition line found, 1201 bp
Strand: minus
Number of exons: 14
Exon 1(-): 207037-207057 (gen) 1-21 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 1 0
Exon 2(-): 205632-205799 (gen) 22-190 (mRNA) id 99.4% mismatches 1 gaps 1 splice site (d a): 1 1
Exon 3(-): 204388-204451 (gen) 191-254 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 1 1
Exon 4(-): 204132-204181 (gen) 255-304 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 1 1
Exon 5(-): 203568-204054 (gen) 305-791 (mRNA) id 96.9% mismatches 15 gaps 0 splice site (d a): 0 1
Exon 6(-): 167094-167110 (gen) 792-808 (mRNA) id 94.1% mismatches 1 gaps 0 splice site (d a): 0 1
Exon 7(-): 164854-164873 (gen) 826-845 (mRNA) id 50.0% mismatches 10 gaps 0 splice site (d a): 1 0
Exon 8(-): 163330-163340 (gen) 846-856 (mRNA) id 72.7% mismatches 3 gaps 0 splice site (d a): 0 1
Exon 9(-): 161658-161670 (gen) 857-869 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 0 1
Exon 10(-): 132199-132222 (gen) 884-907 (mRNA) id 95.8% mismatches 1 gaps 0 splice site (d a): 0 0 uncertain
Exon 11(-): 120008-120023 (gen) 1014-1029 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 0 0 uncertain
Exon 12(-): 102092-102108 (gen) 1102-1118 (mRNA) id 94.1% mismatches 1 gaps 0 splice site (d a): 0 0 uncertain
Exon 13(-): 78872-78885 (gen) 1119-1132 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 0 1
Exon 14(-): 38502-38515 (gen) 1149-1162 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 0 0
Number of splice sites: 5
mRNA coverage: 78%
overall percent identity: 96.6%
Missing mRNA ends: right
Genomic: lcl|SCAF14367 No definition line found
mRNA: lcl|FD0ADA46CC04.contig No definition line found
Exon 1: 207057-207037 (gen) 1-21 (mRNA)
TCTCCTGTGGCTTCTTTCTCTTCTTAAAACGGTGAGTTTTT
|||||||||||||||||||||
CTTCTTTCTCTTCTTAAAACG
N
Exon 2: 205799-205632 (gen) 22-190 (mRNA)
TCTCCTCCAGCTGTCAGCGTATGGCGCCGAGCTGTCATCGGAGGCCTGCAGGGAGCTGGG
||||||||||||||||||||||||||||||||||||||||||||||||||
CTGTCAGCGTATGGCGCCGAGCTGTCATCGGAGGCCTGCAGGGAGCTGGG
A V S V W R R A V I G G L Q G A G
CTTCTCCAGCAACCTGCTGTGCACCTCGTGCGACCTGCTCGGAGAGTTCAGCCTCGCCAA
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CTTCTCCAGCAACCTGCTGTGCACCTCGTGCGACCTGCTCGGAGAGTTCAGCCTCGCCAA
L L Q Q P A V H L V R P A R R V Q P R Q
GCTCCAGCCCAACTGCCAGCAGTGCTGTCAGCAGGAGGCCCTGATGGAAG-GACGCAAGGTAAGGCTCC
|||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||
GCTCCAGCCCAACTGCCAGCAGTGCTGTCAGCAGGAGGCCCTGATGGAAGCGACGCAAG
A P A Q L P A V L S A G G P D G S D A S
Exon 3: 204451-204388 (gen) 191-254 (mRNA)
CTGCACGTAGCTGTATCCCGGGGCCATTCTGGAGGTGTGCGGATGAAAACTGGGGAGGTT
||||||||||||||||||||||||||||||||||||||||||||||||||
CTGTATCCCGGGGCCATTCTGGAGGTGTGCGGATGAAAACTGGGGAGGTT
C I P G P F W R C A D E N W G G
CCCCCAAGTCCAAGGTGAGCCAGA
||||||||||||||
CCCCCAAGTCCAAG
S P K S K
Exon 4: 204181-204132 (gen) 255-304 (mRNA)
ACCTCTTTAGCTTTTGTCAGGAGTGAGAAGCCAAAGATGTTCAAGGGTCTTCAGATCAAGGTAAGAGAGC
||||||||||||||||||||||||||||||||||||||||||||||||||
CTTTTGTCAGGAGTGAGAAGCCAAAGATGTTCAAGGGTCTTCAGATCAAG
L L S G V R S Q R C S R V F R S S
Exon 5: 204054-203568 (gen) 305-791 (mRNA)
ACCCCACCAGTACGTCAGAGGCTCAGACCCTGTGCTCAAGCTTCTGGACGACGAAGGGAA
||||||||||||||||||||||||||||||||||||||||||| ||||||
TACGTCAGAGGCTCAGACCCTGTGCTCAAGCTTCTGGACGACGTAGGGAA
T S E A Q T L C S S F W T T *
CATTGCTGAGGAGCTCAGCATCCTCAAGTGGAACACGGACAGCGTGGAGGAGTTCCTGAG
|||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||
CATTGCTGAGGAACTCAGCATCCTCAAGTGGAACACGGACAGCGTGGAGGAGTTCCTGAG
CGAGAAATTAGATCGGATATAAACGTCAGAATCCGACTTCCCCTTTTTAAACCATCTTCA
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CGAGAAATTAGATCGGATATAAACGTCAGAATCCGACTTCCCCTTTTTAAACCATCTTCG
CTTTCCTTTATTTTTGAGTTATTGTGGGAATCTTCATCCATCACCTGTGTTCAGTGGCTA
|||||||||||||||| ||||||||||||||||||||||| |||||||||||||||||||
CTTTCCTTTATTTTTGCGTTATTGTGGGAATCTTCATCCAACACCTGTGTTCAGTGGCTA
TGAAAATATCTTTTTAAGTTTGCAAGTTTAGTCAGACAGGACTTTCTTTTTGTACTAACA
||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||
TGAAAATATCTTTTTAAGTTTGCAAGTTTAGTCAGACAGGACTCTCTTTTTGTACTAACA
GCGTCCTTGTTGCTCTCCAAAAGCAGATGTCTTGGAAACCCACGTTAGTGAAGACCTGGC
|||||| |||||||||||||||||||||||||||||||||| ||||||||||||||||||
GCGTCCCTGTTGCTCTCCAAAAGCAGATGTCTTGGAAACCCTCGTTAGTGAAGACCTGGC
AGTAAACCAGCGCTGGGATGTGAGGTCTGATGTATATTTGTGGGGTTTTTTCTTCTTCTT
|||||||||||||||||||||||||||||||||||||||||| || |||| ||||||||
AGTAAACCAGCGCTGGGATGTGAGGTCTGATGTATATTTGTGAGGGGTTTTTTTCTTCTT
GTGTTTGGAAAGAAACTGGATTGGTATGTGGGAGTTAAGAACCTGTTTAAGTTTGAAACA
|||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||
GTGTTTGGAAAGAAACTGGATTGGTATGTGGGCGTTAAGAACCTGTTTAAGTTTGAAACA
ATAAACGGAGTCTCTGGAGGTTTCTGT
|||||||| |||||||
ATAAACGGGATCTCTGG
Exon 6: 167110-167094 (gen) 792-808 (mRNA)
AAAATACTAGGAAAAAAAAAAGAAAAACATTCCCAAG
||||||||||| |||||
GAAAAAAAAAAAAAAAA
Exon 7: 164873-164854 (gen) 826-845 (mRNA)
AGAAGCGAGTGGGTGGGGAGTTTCGGCGCTGCGGTAATCA
|||||||| | |
GGGTGGGGGGCCCTGCGCTA
Exon 8: 163340-163330 (gen) 846-856 (mRNA)
TGTACAATTAAACTAAATTGATAAACGTTTT
|||||| | |
AACTAATCTAA
Exon 9: 161670-161658 (gen) 857-869 (mRNA)
ATTATCGAAAAAAAAAAACCTCCATGAATGTCA
|||||||||||||
AAAAAAAACCTCC
Exon 10: 132222-132199 (gen) 884-907 (mRNA)
ATTAACGGTCTCTTTACAAAAAAAAAAAAAAAAAGCCGACATTC
|||||| |||||||||||||||||
TCTTTAAAAAAAAAAAAAAAAAAA
Exon 11: 120023-120008 (gen) 1014-1029 (mRNA)
CGTCCCACCATTTTTTTTTTCAAATTGCTGCAGTCA
||||||||||||||||
TTTTTTTTTTCAAATT
Exon 12: 102108-102092 (gen) 1102-1118 (mRNA)
AGATTTTAGCTAAAAAATAAAAAAGCAGGAGCCGTCG
||||||| |||||||||
TAAAAAAAAAAAAAGCA
Exon 13: 78885-78872 (gen) 1119-1132 (mRNA)
CACTGCTGGAAAAACAAAAAAAAAGGCTGGATAA
||||||||||||||
AAAACAAAAAAAAA
Exon 14: 38515-38502 (gen) 1149-1162 (mRNA)
TCCAAACAGCATCAAAAAAAAAAATGCATTTAAA
||||||||||||||
ATCAAAAAAAAAAA
--SPIDEY version 1.40--
Genomic: lcl|SCAF14367 No definition line found, 444931 bp
mRNA: lcl|FD0ADA50AE12.contig No definition line found, 977 bp
No alignment found.
--SPIDEY version 1.40--
Genomic: lcl|SCAF14367 No definition line found, 444931 bp
mRNA: lcl|FD0AEB7DG04.contig No definition line found, 936 bp
No alignment found.
--SPIDEY version 1.40--
Genomic: lcl|SCAF14367 No definition line found, 444931 bp
mRNA: lcl|FD0AEB22AD04.contig No definition line found, 858 bp
Strand: minus
Number of exons: 6
Exon 1(-): 207037-207095 (gen) 1-59 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 1 0
Exon 2(-): 205632-205799 (gen) 60-227 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 1 1
Exon 3(-): 204388-204451 (gen) 228-291 (mRNA) id 98.4% mismatches 1 gaps 0 splice site (d a): 1 1
Exon 4(-): 204132-204181 (gen) 292-341 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 1 1
Exon 5(-): 203568-204054 (gen) 342-828 (mRNA) id 99.2% mismatches 4 gaps 2 splice site (d a): 0 1
Exon 6(-): 133951-133980 (gen) 829-858 (mRNA) id 60.0% mismatches 12 gaps 0 splice site (d a): 0 1
Number of splice sites: 4
mRNA coverage: 100%
overall percent identity: 98.0%
Missing mRNA ends: neither
Genomic: lcl|SCAF14367 No definition line found
mRNA: lcl|FD0AEB22AD04.contig No definition line found
Exon 1: 207095-207037 (gen) 1-59 (mRNA)
TGGTTTGTTGCCAGCTCCAAAATGTCGGGAGAGGTTTATCTCCTGTGGCTTCTTTCTCTT
||||||||||||||||||||||||||||||||||||||||||||||||||
CCAGCTCCAAAATGTCGGGAGAGGTTTATCTCCTGTGGCTTCTTTCTCTT
S S K M S G E V Y L L W L L S L
CTTAAAACGGTGAGTTTTT
|||||||||
CTTAAAACG
L K T
Exon 2: 205799-205632 (gen) 60-227 (mRNA)
TCTCCTCCAGCTGTCAGCGTATGGCGCCGAGCTGTCATCGGAGGCCTGCAGGGAGCTGGG
||||||||||||||||||||||||||||||||||||||||||||||||||
CTGTCAGCGTATGGCGCCGAGCTGTCATCGGAGGCCTGCAGGGAGCTGGG
L S A Y G A E L S S E A C R E L G
CTTCTCCAGCAACCTGCTGTGCACCTCGTGCGACCTGCTCGGAGAGTTCAGCCTCGCCAA
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CTTCTCCAGCAACCTGCTGTGCACCTCGTGCGACCTGCTCGGAGAGTTCAGCCTCGCCAA
F S S N L L C T S C D L L G E F S L A K
GCTCCAGCCCAACTGCCAGCAGTGCTGTCAGCAGGAGGCCCTGATGGAAGGACGCAAGGT
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
GCTCCAGCCCAACTGCCAGCAGTGCTGTCAGCAGGAGGCCCTGATGGAAGGACGCAAG
L Q P N C Q Q C C Q Q E A L M E G R K
AAGGCTCC
Exon 3: 204451-204388 (gen) 228-291 (mRNA)
CTGCACGTAGCTGTATCCCGGGGCCATTCTGGAGGTGTGCGGATGAAAACTGGGGAGGTT
||||| ||||||||||||||||||||||||||||||||||||||||||||
CTGTACCCCGGGGCCATTCTGGAGGTGTGCGGATGAAAACTGGGGAGGTT
L Y P G A I L E V C G *
CCCCCAAGTCCAAGGTGAGCCAGA
||||||||||||||
CCCCCAAGTCCAAG
Exon 4: 204181-204132 (gen) 292-341 (mRNA)
ACCTCTTTAGCTTTTGTCAGGAGTGAGAAGCCAAAGATGTTCAAGGGTCTTCAGATCAAGGTAAGAGAGC
||||||||||||||||||||||||||||||||||||||||||||||||||
CTTTTGTCAGGAGTGAGAAGCCAAAGATGTTCAAGGGTCTTCAGATCAAG
Exon 5: 204054-203568 (gen) 342-828 (mRNA)
ACCCCACCAGTACGTCAGAGGCTCAGACCCTGTGCTCAAGCTTCTGGACGACGAAGGGAA
||||||||||||||||||||||||||||||||||||||||||||||||||
TACGTCAGAGGCTCAGACCCTGTGCTCAAGCTTCTGGACGACGAAGGGAA
CATTGCTGAGGAGCTCAGCATCCTCAAGTGGAACACGGACAGCGTGGAGGAGTTCCTGAG
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CATTGCTGAGGAGCTCAGCATCCTCAAGTGGAACACGGACAGCGTGGAGGAGTTCCTGAG
CGAGAAATTAGATCGGATATAAACGTCAGAATCCGACTTCCCCTTTTTAAACCATCTTCA
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CGAGAAATTAGATCGGATATAAACGTCAGAATCCGACTTCCCCTTTTTAAACCATCTTCA
CTTTCCTTTATTTTTGAGTTATTGTGGGAATCTTCATCCATCACCTGTGTTCAGTGGCTA
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CTTTCCTTTATTTTTGAGTTATTGTGGGAATCTTCATCCATCACCTGTGTTCAGTGGCTA
TGAAAATATCTTTTTAAGTTTGCAAGTTTAGTCAGACAGGACTTTCTTTTTGTACTAACA
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
TGAAAATATCTTTTTAAGTTTGCAAGTTTAGTCAGACAGGACTTTCTTTTTGTACTAACA
GCGTCCTTGTTGCTCTCCAAAAGCAGATGTCTTGGAAACCCACGTTAGTGAAGACCTGGC
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
GCGTCCTTGTTGCTCTCCAAAAGCAGATGTCTTGGAAACCCACGTTAGTGAAGACCTGGC
AGTAAACCAGCGCTGGGATGTGAGGTCTGATGTATATTTGTGGGGTTTTTTCTTCTTCTT
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
AGTAAACCAGCGCTGGGATGTGAGGTCTGATGTATATTTGTGGGGTTTTTTCTTCTTCTT
GTGTTTGGAAAGAAACTGGATTGGTATGT-GGGAGTTAAGAACCTGTTTAAGTTTGAAAC
||| ||||||||||||||||||||||||| ||||||||||||||||||||||||||||||
GTG-TTGGAAAGAAACTGGATTGGTATGTGGGGAGTTAAGAACCTGTTTAAGTTTGAAAC
AATAAACGGAGTCTCTGGAGGTTTCTGT
||||||||||||||| |
AATAAACGGAGTCTCGGA
Exon 6: 133980-133951 (gen) 829-858 (mRNA)
TGTCACGAGAAAAAAAAAAACAAAAAAACTCCATTCATAGCTCGAGCAAC
|||||||||| ||||||| |
AAAAAAAAAAAAAAAAAAGGCNCACATGGC
--SPIDEY version 1.40--
Genomic: lcl|SCAF14367 No definition line found, 444931 bp
mRNA: lcl|FD0AFB9BC06.contig No definition line found, 885 bp
Strand: minus
Number of exons: 5
Exon 1(-): 207034-207130 (gen) 1-97 (mRNA) id 99.0% mismatches 1 gaps 0 splice site (d a): 0 0
Exon 2(-): 205632-205796 (gen) 98-262 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 1 1
Exon 3(-): 204388-204451 (gen) 263-326 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 1 1
Exon 4(-): 204132-204181 (gen) 327-376 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 1 1
Exon 5(-): 203567-204054 (gen) 377-866 (mRNA) id 98.2% mismatches 9 gaps 4 splice site (d a): 0 1
Number of splice sites: 3
mRNA coverage: 100%
overall percent identity: 98.8%
Missing mRNA ends: neither
Non-aligning poly(A)+ tail length: 20
Genomic: lcl|SCAF14367 No definition line found
mRNA: lcl|FD0AFB9BC06.contig No definition line found
Exon 1: 207130-207034 (gen) 1-97 (mRNA)
GGTTTCTCTCAAAGTAAAAGAAGAAGAGGGGACGGTGGTTTGTTGCCAGCTCCAAAATGT
||||||||||||||||||||||||||||||||||||||||||||||||||
AAAGTAAAAGAAGAAGAGGGGACGGTGGTTTGTTGCCAGCTCCAAAATGT
K K K R G R W F V A S S K M
CGGGAGAGGTTTATCTCCTGTGGCTTCTTTCTCTTCTTAAAACGGTGAGTTTTTTTT
|||||||||||||||||||||||||||||||||||||||||||| ||
CGGGAGAGGTTTATCTCCTGTGGCTTCTTTCTCTTCTTAAAACGTTG
S G E V Y L L W L L S L L K T L
Exon 2: 205796-205632 (gen) 98-262 (mRNA)
CCTCCAGCTGTCAGCGTATGGCGCCGAGCTGTCATCGGAGGCCTGCAGGGAGCTGGGCTT
||||||||||||||||||||||||||||||||||||||||||||||||||
TCAGCGTATGGCGCCGAGCTGTCATCGGAGGCCTGCAGGGAGCTGGGCTT
S A Y G A E L S S E A C R E L G F
CTCCAGCAACCTGCTGTGCACCTCGTGCGACCTGCTCGGAGAGTTCAGCCTCGCCAAGCT
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CTCCAGCAACCTGCTGTGCACCTCGTGCGACCTGCTCGGAGAGTTCAGCCTCGCCAAGCT
S S N L L C T S C D L L G E F S L A K L
CCAGCCCAACTGCCAGCAGTGCTGTCAGCAGGAGGCCCTGATGGAAGGACGCAAGGTAAG
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
CCAGCCCAACTGCCAGCAGTGCTGTCAGCAGGAGGCCCTGATGGAAGGACGCAAG
Q P N C Q Q C C Q Q E A L M E G R K
GCTCC
Exon 3: 204451-204388 (gen) 263-326 (mRNA)
CTGCACGTAGCTGTATCCCGGGGCCATTCTGGAGGTGTGCGGATGAAAACTGGGGAGGTT
||||||||||||||||||||||||||||||||||||||||||||||||||
CTGTATCCCGGGGCCATTCTGGAGGTGTGCGGATGAAAACTGGGGAGGTT
L Y P G A I L E V C G *
CCCCCAAGTCCAAGGTGAGCCAGA
||||||||||||||
CCCCCAAGTCCAAG
Exon 4: 204181-204132 (gen) 327-376 (mRNA)
ACCTCTTTAGCTTTTGTCAGGAGTGAGAAGCCAAAGATGTTCAAGGGTCTTCAGATCAAGGTAAGAGAGC
||||||||||||||||||||||||||||||||||||||||||||||||||
CTTTTGTCAGGAGTGAGAAGCCAAAGATGTTCAAGGGTCTTCAGATCAAG
Exon 5: 204054-203567 (gen) 377-866 (mRNA)
ACCCCACCAGTACGTCAGAGGCTCAGACCCTGTGCTCAAGCTTCTGGACGACGAAGGGAA
||||||||||||||||||||||||||||||||||||||||||||||||||
TACGTCAGAGGCTCAGACCCTGTGCTCAAGCTTCTGGACGACGAAGGGAA
CATTGCTGAGGAGCTCAGCATCCTCAAGTGGAACACGGACAGCGTGGAGGAGTTCCTGAG
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CATTGCTGAGGAGCTCAGCATCCTCAAGTGGAACACGGACAGCGTGGAGGAGTTCCTGAG
CGAGAAATTAGATCGGATATAAACGTCAGAATCCGACTTCCCCTTTTTAAACCATCTTCA
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CGAGAAATTAGATCGGATATAAACGTCAGAATCCGACTTCCCCTTTTTAAACCATCTTCA
CTTTCCTTTATTTTTGAGTTATTGTGGGAATCTTCATCCATCACCTGTGTTCAGTGGCTA
|||||||||||||||| ||||||||||||||||||||||| |||||||||||||||||||
CTTTCCTTTATTTTTGCGTTATTGTGGGAATCTTCATCCAACACCTGTGTTCAGTGGCTA
TGAAAATATCTTTTTAAGTTTGCAAGTTTAGTCAGACAGGAC---TTTCTTTTTGTACTA
|||||||||||||||||||||||||||||||||||||||||| |||||||||||||||
TGAAAATATCTTTTTAAGTTTGCAAGTTTAGTCAGACAGGACTATTTTCTTTTTGTACTA
ACAGCGTCCTTGTTGCTCTCCAAAAGCAGATGTCTTGGAAACCCACGTTAGTGAAGACCT
|||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||
ACAGCGTCCTTGTTGCTCTCCAAAAGCAGATGTCTTGGAAACCCTCGTTAGTGAAGACCT
GGCAGTAAACCAGCGCTGGGATGTGAGGTCTGATGTATATTTGTGGGGTTTTTTCTTCTT
|||||||||||||||||||||||||||||||||||||||||||||||| ||||| |||||
GGCAGTAAACCAGCGCTGGGATGTGAGGTCTGATGTATATTTGTGGGGGTTTTT-TTCTT
CTTGTGTTTGGAAAGAAACTGGATTGGTATGTGGGAGTTAAGAACCTGTTTAAGTTTGAA
||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||
CTTGTGTTTGGAAAGAAACTGGATTGGTATGTGGGCGTTAAGAACCTGTTTAAGTTTGAA
ACAATAAACGGAGTCTCTGGAGGTTTCTGTG
|||||||||||||||||||||
ACAATAAACGGAGTCTCTGGA
--SPIDEY version 1.40--
Genomic: lcl|SCAF14367 No definition line found, 444931 bp
mRNA: lcl|FD0AHA40CG09.contig No definition line found, 1418 bp
Strand: minus
Number of exons: 5
Exon 1(-): 207037-207125 (gen) 575-663 (mRNA) id 100.0% mismatches 0 gaps 0 splice site (d a): 1 0
Exon 2(-): 205632-205799 (gen) 664-831 (mRNA) id 98.2% mismatches 3 gaps 0 splice site (d a): 1 1
Exon 3(-): 204388-204451 (gen) 832-895 (mRNA) id 98.4% mismatches 1 gaps 0 splice site (d a): 1 1
Exon 4(-): 204132-204181 (gen) 896-945 (mRNA) id 94.0% mismatches 3 gaps 0 splice site (d a): 1 1
AATGAAATTATTATTATTTTTTTTATTTTGGTA
||||| |||||||
TTATTTTTTTTTT
Exon 11: 196200-196193 (gen) 1051-1058 (mRNA)
TCCCGCTTTTCTCCCCTTCTCTGGCTTC
||||||||
CTCCCCTT
Exon 12: 195567-195563 (gen) 1059-1063 (mRNA)
AATGCTGCTTTTCCACATCGAATGT
|||||
TTCCA
Exon 13: 195513-195496 (gen) 1080-1097 (mRNA)
ATTTTCTGCTTTCTTTTTTTTTTTTTTTAGATGCTTTT
||||||||||||||||||
TTCTTTTTTTTTTTTTTT

1 FD0ABA26AC03.contig: 988 1086 upstemen:-11.79 fullstructen: -27.54 GGAAUUGUUU UCUACCU GCUGCGAACCUCA AUGAA GCCUCACGCuG aA AAUCCGCC AGGCCUGAGGU CGAU GCUCACUGUU CCGGUGGg ucAAACAAAC
Els contigs de cDNA i ESTs que contenen l'element SECIS són:
BLASTN 2.2.5 [Nov-16-2002]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= FD0ABA26AC03.contig
(1123 letters)
Database: blast/hola
1 sequences; 444,931 total letters
Searching.done
Score E
Sequences producing significant alignments: (bits) Value
SCAF14367 30 0.39
>SCAF14367
Length = 444931
Score = 30.2 bits (15), Expect = 0.39
Identities = 15/15 (100%)
Strand = Plus / Plus
Query: 773 ctccaacatgaccac 787
|||||||||||||||
Sbjct: 204347 ctccaacatgaccac 204361
Score = 30.2 bits (15), Expect = 0.39
Identities = 15/15 (100%)
Strand = Plus / Plus
Query: 194 cctctcctctcaggc 208
|||||||||||||||
Sbjct: 124414 cctctcctctcaggc 124428
Score = 30.2 bits (15), Expect = 0.39
Identities = 15/15 (100%)
Strand = Plus / Minus
Query: 1094 atggttcaagtggaa 1108
|||||||||||||||
Sbjct: 251227 atggttcaagtggaa 251213
Score = 30.2 bits (15), Expect = 0.39
Identities = 15/15 (100%)
Strand = Plus / Minus
Query: 818 gttaagccaggcccg 832
|||||||||||||||
Sbjct: 216861 gttaagccaggcccg 216847
Score = 30.2 bits (15), Expect = 0.39
Identities = 15/15 (100%)
Strand = Plus / Plus
Query: 790 gattcctgaggtgag 804
|||||||||||||||
Sbjct: 243033 gattcctgaggtgag 243047
Score = 30.2 bits (15), Expect = 0.39
Identities = 15/15 (100%)
Strand = Plus / Plus
Query: 879 tcatcatcagtcatt 893
|||||||||||||||
Sbjct: 194668 tcatcatcagtcatt 194682
Score = 28.2 bits (14), Expect = 1.6
Identities = 14/14 (100%)
Strand = Plus / Minus
Query: 346 agcccgagtggaga 359
||||||||||||||
Sbjct: 145447 agcccgagtggaga 145434
Score = 28.2 bits (14), Expect = 1.6
Identities = 17/18 (94%)
Strand = Plus / Minus
Query: 1077 tcaaacaaacacacagga 1094
||||| ||||||||||||
Sbjct: 183028 tcaaaaaaacacacagga 183011
Score = 28.2 bits (14), Expect = 1.6
Identities = 17/18 (94%)
Strand = Plus / Plus
Query: 898 atgtgtcctttttgtgaa 915
||||||||||||| ||||
Sbjct: 225034 atgtgtcctttttatgaa 225051
Score = 28.2 bits (14), Expect = 1.6
Identities = 14/14 (100%)
Strand = Plus / Minus
Query: 302 tcctccttccaggc 315
||||||||||||||
Sbjct: 118376 tcctccttccaggc 118363
Score = 28.2 bits (14), Expect = 1.6
Identities = 14/14 (100%)
Strand = Plus / Plus
Query: 408 tgacccaggacatt 421
||||||||||||||
Sbjct: 336371 tgacccaggacatt 336384
Score = 28.2 bits (14), Expect = 1.6
Identities = 14/14 (100%)
Strand = Plus / Plus
Query: 903 tcctttttgtgaac 916
||||||||||||||
Sbjct: 90590 tcctttttgtgaac 90603
Score = 28.2 bits (14), Expect = 1.6
Identities = 14/14 (100%)
Strand = Plus / Plus
Query: 269 tggctgatggtgct 282
||||||||||||||
Sbjct: 361601 tggctgatggtgct 361614
Score = 28.2 bits (14), Expect = 1.6
Identities = 14/14 (100%)
Strand = Plus / Minus
Query: 977 tccgtgtgtgtgga 990
||||||||||||||
Sbjct: 181747 tccgtgtgtgtgga 181734
Score = 28.2 bits (14), Expect = 1.6
Identities = 14/14 (100%)
Strand = Plus / Minus
Query: 302 tcctccttccaggc 315
||||||||||||||
Sbjct: 110230 tcctccttccaggc 110217
Score = 28.2 bits (14), Expect = 1.6
Identities = 14/14 (100%)
Strand = Plus / Plus
Query: 348 cccgagtggagagc 361
||||||||||||||
Sbjct: 36452 cccgagtggagagc 36465
Score = 26.3 bits (13), Expect = 6.1
Identities = 16/17 (94%)
Strand = Plus / Minus
Query: 308 ttccaggcggacctgca 324
|||| ||||||||||||
Sbjct: 275065 ttccgggcggacctgca 275049
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 1107 aagtgtttaaaaa 1119
|||||||||||||
Sbjct: 377837 aagtgtttaaaaa 377849
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 575 aagagggctcaac 587
|||||||||||||
Sbjct: 274629 aagagggctcaac 274641
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 358 gagctgtggagga 370
|||||||||||||
Sbjct: 1329 gagctgtggagga 1317
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 192 tccctctcctctc 204
|||||||||||||
Sbjct: 55998 tccctctcctctc 55986
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 254 cagcctggaacca 266
|||||||||||||
Sbjct: 23548 cagcctggaacca 23536
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 666 cctccacctcctc 678
|||||||||||||
Sbjct: 214439 cctccacctcctc 214451
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 951 tcccgattctcac 963
|||||||||||||
Sbjct: 239696 tcccgattctcac 239708
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 710 gaaacagaagaga 722
|||||||||||||
Sbjct: 427381 gaaacagaagaga 427393
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 981 tgtgtgtggaatt 993
|||||||||||||
Sbjct: 105808 tgtgtgtggaatt 105796
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 615 gcttctctcctgc 627
|||||||||||||
Sbjct: 281701 gcttctctcctgc 281713
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 1107 aagtgtttaaaaa 1119
|||||||||||||
Sbjct: 276152 aagtgtttaaaaa 276140
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 450 acattcctggggc 462
|||||||||||||
Sbjct: 443483 acattcctggggc 443471
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 1048 ctgaggtcgatgc 1060
|||||||||||||
Sbjct: 12723 ctgaggtcgatgc 12711
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 879 tcatcatcagtca 891
|||||||||||||
Sbjct: 328395 tcatcatcagtca 328407
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 295 ctgcgtctcctcc 307
|||||||||||||
Sbjct: 33557 ctgcgtctcctcc 33545
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 670 cacctcctctcct 682
|||||||||||||
Sbjct: 401962 cacctcctctcct 401950
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 673 ctcctctcctgtc 685
|||||||||||||
Sbjct: 308742 ctcctctcctgtc 308754
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 264 ccatgtggctgat 276
|||||||||||||
Sbjct: 308548 ccatgtggctgat 308536
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 686 ccagaggacgcct 698
|||||||||||||
Sbjct: 157770 ccagaggacgcct 157782
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 386 ctcagagaggtca 398
|||||||||||||
Sbjct: 281030 ctcagagaggtca 281042
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 41 cggaccagattcc 53
|||||||||||||
Sbjct: 358745 cggaccagattcc 358733
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 204 caggcctgatcca 216
|||||||||||||
Sbjct: 146753 caggcctgatcca 146741
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 323 cagaagctggacg 335
|||||||||||||
Sbjct: 226033 cagaagctggacg 226045
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Minus
Query: 622 tcctgccaaggac 634
|||||||||||||
Sbjct: 163893 tcctgccaaggac 163881
Score = 26.3 bits (13), Expect = 6.1
Identities = 13/13 (100%)
Strand = Plus / Plus
Query: 322 gcagaagctggac 334
|||||||||||||
Sbjct: 93337 gcagaagctggac 93349
Database: blast/hola
Posted date: Mar 10, 2004 7:01 PM
Number of letters in database: 444,931
Number of sequences in database: 1
Lambda K H
1.37 0.711 1.31
Gapped
Lambda K H
1.37 0.711 1.31
Matrix: blastn matrix:1 -3
Gap Penalties: Existence: 5, Extension: 2
Number of Hits to DB: 354
Number of Sequences: 1
Number of extensions: 354
Number of successful extensions: 42
Number of sequences better than 10.0: 1
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 42
length of query: 1123
length of database: 444,931
effective HSP length: 14
effective length of query: 1109
effective length of database: 444,917
effective search space: 493412953
effective search space used: 493412953
T: 0
A: 0
X1: 6 (11.9 bits)
X2: 15 (29.7 bits)
S1: 12 (24.3 bits)
S2: 13 (26.3 bits)