Iron Response ElementS
![]() METHODS, PROCEDURE
AND RESULTS
METHODS, PROCEDURE
AND RESULTS
![]()
![]() ASSESSING
THERMODYNAMIC STABILITY
ASSESSING
THERMODYNAMIC STABILITY
In order to determine how stable IREs strucutures were, we used a program called RNAfold (http://bioweb.pasteur.fr/seqanal/interfaces/rnafold-simple.html) that calculates the minimum free energy of the thermodynamic ensemble mRNAs structures. As the program uses mRNA as template, the genetic code of the sequences introduced has to be in the correct form. It was necessary then to change all the timidines present in the sequences to the uracile residues. The commands used at Perl console to do it can be seen if click here.
We did a raw prediction of the thermodynamic stability because after doing a constrained one we realize that no importance differences were observed. The constrained prediction was not very useful because even though making our pattern more general than originally, the result of RNAfold was similar in both cases.
RNAfold show us the optimal secondary structure symbolized with brackets and points, and it also shows the minimum free energy in kcal/mol of this structure. The more negative RNAfold results is, the more stable thermodynamically the structure is.
The secondary structure predicted by RNAfold is due to the pattern introduced when the is program run. This way we could check if all the predicted structures corresponded to the real IREs structure:
- Ferritin_training set: we obtained not very high results (understood as not very negative) because to build the pattern we had only used one nucleotide upstream the loop-bulge. For this reason, the secondary structre was not as stable as if more base pairings had been included in the steem while building the pattern. Two of these results have higher values than the others because the minimum free energy structure calculated by RNAfold is not the real IREs structure.
- Ferrintin_test set: the same as before the results obtained are not very negative and the reason is the same one, we just took one nucleotide upstream the middle loop to construct the pattern. As told if more nucleotides were used higher (more negative) the energy would have been. One of these results has a higher value than the others because the minimum free energy structure calculated by RNAfold is not the real IREs structure.
- Transferrin_training set : we obtained high negative results because after the C-bulge there were many base pairs and it made its structure more stable. In this case the minimum free energy secondary structure predicted corresponds to the real IREs structure.
- Transferrin_test set: once we run RNAfold over these sequences we obtained the same as with the transferring training set really high results. In this occasion the sequences from which the patter was built a great deal of number of base pairs after the C-bulge.
- Aconitase: we obtained not pretty negative because upsteam the C-bulge we have not used a lot of nucleotides so as consequence the secondary structure obtained is not very good, the same that happened with other sequences checked.
- Ferroportin: high results (negative) were obtained because after the C-bulge there were quite enough base pairs for making the structure stable. The minimum free energy secondary structure predicted corresponds to the real IREs structure.
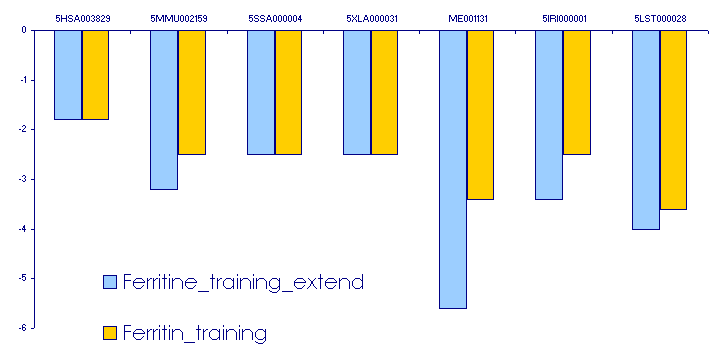
After assessing the thermodynamic stability of all these structures (and observing that some of them were not very stable) we wanted to check if increasing the lenght of our ferritin sequences, the results obtained would be the same or it would be more constrained.
We repeated all the processes made before but using the sequences elongated:
- At first we used for running the Patscan over the elongated set the ferritin pattern, but as it was pretty strict (pattern not shown) we got the initial ferritin sequences but without any elongation. For this reason we used a more general pattern obtained from EMBL database (a general pattern for all the training sequences) with which all the ferritin elongated sequences were recognized.
- With the new results RNAfold was run. As expected there were some differences between the short and long ferritin sequences. We can observe that with the long sequences the thermodynamic stability increases (RNAfold gives more negative values than with the initial ferritin training set). The nucleotides added in ferritin sequences allow a more stable secondary structure of IREs (the steem is longer than before).
We have represented the differences in the following: