Iron Response ElementS
![]() STATE OF ART
STATE OF ART
![]()
![]() IRON REGULATION PATHWAY
IRON REGULATION PATHWAY
Many circumstances including alterations in iron availability, the level of specific hormones, growth factors and cytokines, as well as the rate of cell proliferation or differentiation, influence the uptake and metabolic use of iron. The size of the intracellular chelatable iron pool influences ferritin and TfR gene expression largely at the post-transcriptional level through the action of two iron-regulated RNA binding proteins, the IRPs. Elucidation of the central role of IRPs, and the iron responsive elements (IREs) to which they bind, in regulating ferrintin and TfR synthesis has been the key in elucidating how iron-dependent and iron-independent signaling pathways influence cellular iron homeostasis.
The translation of some mRNA molecules is blocked by specific translation repressor proteins that bind near the 5' end of the mRNAs where translation would begin. This kind of mechanism is called negative translation control.
When intracellular stores of iron are low due to a poor envirorment, the ferritin function is not required because if the low iron ions present in the cell have to be used to mantain the correct cell metabolism it is not logic that ferritin keeps on accumullating. So, this is the point where ferritin mRNAs translation has to be blocked. IRPs binds to the ferritin 5' IRE inhibiting translation initiation by blocking the ability of the 40S ribosomal subunit to scan for the AUG start codon where it initially binds. On the opposite hand, when iron concentrations are high, IREs binding proteins does not bind to the 5' ferritin IRE so that translation iniciation can proceed and increase the protein pool.
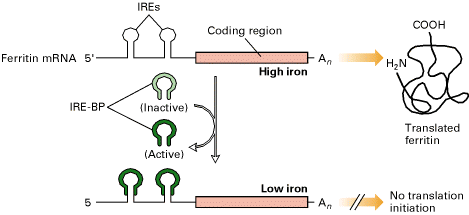
We have to say that ferritin mRNA masked or binded to IRP is not destroyed, just is released into the cell waiting for conditions to be suitable again.
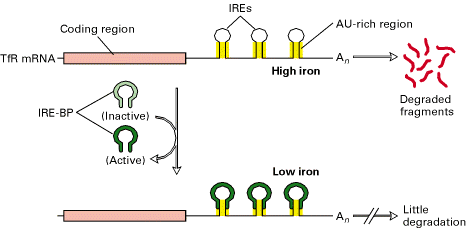
Many other RNAs are unstable because they contain specific sequences that stimulate their degradation. A long sequence rich in A and U nucleotides in the 3' UTR cause them to be unstable. This AU-rich sequence appears to accelerate mRNA degradation by stimulating the removal of the poly-A tail found at the 3' end of almost all eukaryotic mRNAs. Other unstable mRNAs contain recognition sites in their 3' UTR for specific endonucleases that cleave the mRNA but that have exactly the same consequence, a decrease in mRNA concentration and the protein encoded.
The stability of an mRNA can be changed in response to extracellular signals. Addition of iron to cells decreases the stability of the mRNA that encodes the receptor protein that binds the iron-transporting protein transferrin, causing less of this receptor to be made. In this case the stability is modulated by IRPs and the five IRE structures present at the 3' UTR.
In the transferrin receptor mRNA stability depends on the five IREs in the 3' untranslated region of the mRNA and on IRPs, whose conformation differs as told at high and low iron levels. Each IRE forms a stem-loop structure in which the stem contains AU-rich sequences similar to those that destabilize mRNAs inducing its degradation. When iron concentrations falls slightly the cell increases the need to uptake iron from the exterior to maintain physiological concentrations and then the functions mediated by it. The IRP binding to the IREs is thought to block recognition of the destabilizing AU-rich sequences by the proteins that would otherwise degrade transferrin receptor mRNA. In contrast, high iron concentration would result detrimental to cell because of its toxic nature, so transferrin receptor should reduce its activity. In this occasion, IREs at 3' region keeps naked (not IRP binded) so AU-rich sequences are recognized by endonucleases and mRNA is degraded.