Iron Response ElementS
![]() STATE OF ART
STATE OF ART
![]()
![]() IREs STRUCTURE
IREs STRUCTURE
IREs have been identified in a great range of organisms (vertebrates,
invertebrates and bacteria but not in plants) and as seen recently iron and
oxygen metabolisms proteins are not the only ones that carry this structural
motif in their structure. Many heat shock housekeeping proteins, proto-oncogenes,
chlorophyll-binding proteins and ribonucleotide reductases also keep them in their messenger.
Originally it was thought all IREs, 5' and 3', were structurally the same because of the predicted secondary structure. However, comparisons
among larger numbers of IREs sequences from different mRNAs have shown that
there are specific nucleotide changes that would confer to the mRNAs different
regulation due to the different interaction with IRPs. Then, many IREs isoforms
can be defined up the nucleotide pattern presented.
- IREs
primary structure
: Comparison of animal IRE sequences reveals that the conservation of sequence
identity is much higher between species for the same mRNA than between different
mRNAs in the same species (90% vs 36-85%). All of them but, are formed by
between 26 and 30 nucleotides in which a central CAGWUN
nucleotide sequence and a C residue
five bases upstream are totaly conserved. In ferritin appears an additional set
of conserved bases MGC that as we will see later has a special function in the
regulation pathway.
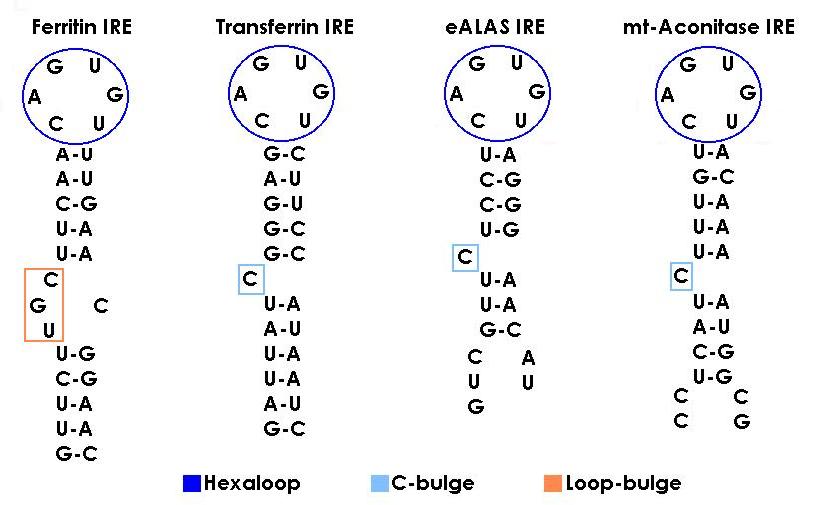
- IREs secondary structure : The CAGWGX sequence folds in a terminal hexaloop in which C1 and G5 form a base pair required for IRP binding. It has been seen that a substitution of this C-G for A-U prevents binding of either IRP-1 or IRP-2. The conformational pattern that provides the C-G alignment to the IRE is probably crucial for the RNA-protein interaction. Under the hexaloop the IRE contains a stem of 9 to 10 base pairs that fold in a a-helix which is always interrupted by either the C residue mencioned before, known as C-bulge (in transferrin from iron metabolism and eALAS and mt-aconitase from the oxygen one), or the MGC sequence called as well loop bulge (just present in ferritin mRNA).
Recent experiments have determined through directed mutagenesis that the residues between the hexaloop and the C-bulge or loop bulge, also contribute to the protein binding, so not all variations in sequence are allowed.
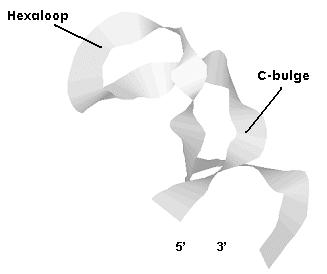
The structure showed is from the transferrin protein. Remember that the C-bulge would be
substituted by the loop-bulge in the rest of IREs containing mRNAs.
- IREs tertiary structure: NMR spectroscopy has shown the C-G base pair across the CAGWGN hexaloop pushes the AGW into the solvent what facilitates IRP-binding with the hairpin. Both, the C-bulge and the loop-bulge enhance due to the distortion introduced in the helix, specific base and ribose contact sites. Moreover the internal ferritin loop-bulge allows metal binding what selectively enhances IRP2 binding. This make us pay attention in which way the primary structure influences how the messenger is going to fold and in consequence how and with who is going to interact.