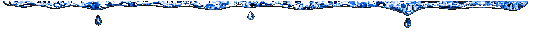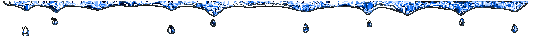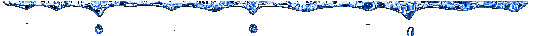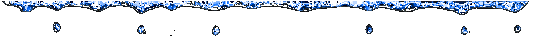†
![]() † EVOLUTION TREES:
† EVOLUTION TREES:
![]() †Neighbor-joining
consens tree:
†Neighbor-joining
consens tree:
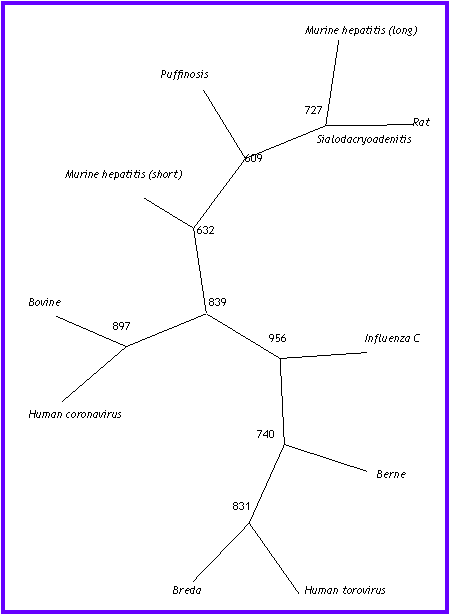 In this evolution tree we can appreciate two big
clusters, which are situated on the top and on the bottom of the tree. They
correspond to the two genus of Coronaviridae family. The cluster that is
on the top belongs to the genus Coronavirus, and the other belongs to
the genus Torovirus.
In this evolution tree we can appreciate two big
clusters, which are situated on the top and on the bottom of the tree. They
correspond to the two genus of Coronaviridae family. The cluster that is
on the top belongs to the genus Coronavirus, and the other belongs to
the genus Torovirus.
Also, inside the Coronavirus
cluster, we can observe two couples: Bovine and human coronavirus, and
murine hepatitis virus and rat sialodacryoadenitis coronavirus.
The similarity of the members of these couples maybe is explained by the differences between their hosts, the first infects superior mammals, and the others rodents.
Besides these big clusters, we found Influenza C branch
that are completely independent of the rest of the sequences. This can be
explained because it belongs a different viruses family.
![]() † Parsimony
consens tree:
† Parsimony
consens tree:
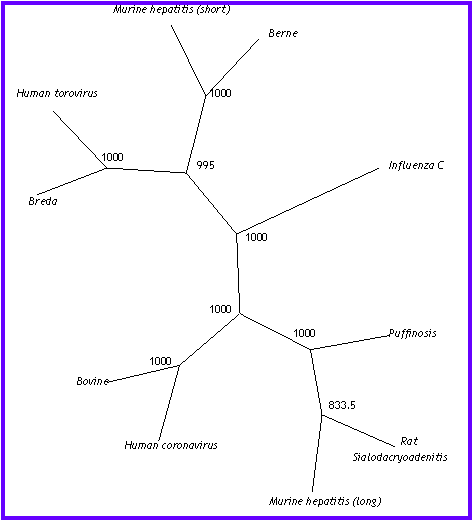 In this tree, appear the same clusters than in
the neighbor-joining analysis, but we can see a difference on the Murine
hepatitis virus (the shortest) position, that changes from Coronavirus
cluster to Torovirus group. †This
branch is few strange, because it puts together viruses that belong at different
genus.
In this tree, appear the same clusters than in
the neighbor-joining analysis, but we can see a difference on the Murine
hepatitis virus (the shortest) position, that changes from Coronavirus
cluster to Torovirus group. †This
branch is few strange, because it puts together viruses that belong at different
genus.
Influenza
continues being independent as we hope after study NJ tree.
![]() † Maxim
likelihood tree:
† Maxim
likelihood tree:
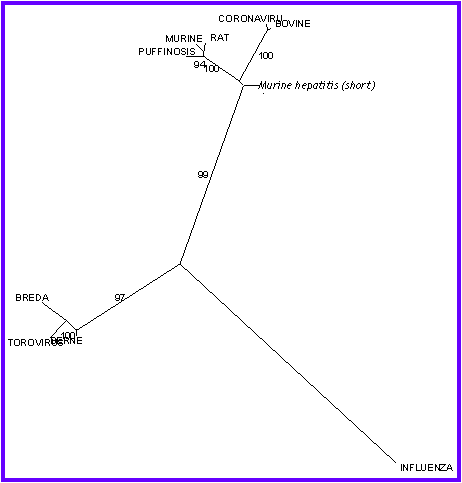 This tree looks very different than the others, but the clusters that we
found are the same, and Influenza continues like an independent branch.
This tree looks very different than the others, but the clusters that we
found are the same, and Influenza continues like an independent branch.
Inside the two clusters, we canít analyze the relation
between the organisms, because the branches are very short.