About
Hystrix brachyura
Hystrix brachyura is a porcupine, believed to have originated in Southern Asia. The aim of this project is to characterize its selenoproteome (all its selenoproteins) as well as the machinery it needs to be translated.
Taxonomy
| Kingdom | Animalia |
|---|---|
| Phylum | Chordata |
| Class | Mammalia |
| Order | Rodentia |
| Family | Hystricidae |
| Genus | Hystrix |
| Subgenus | Hystrix (Acanthion) |
| Species | Hystrix brachyura Linnaeus |
Distribution
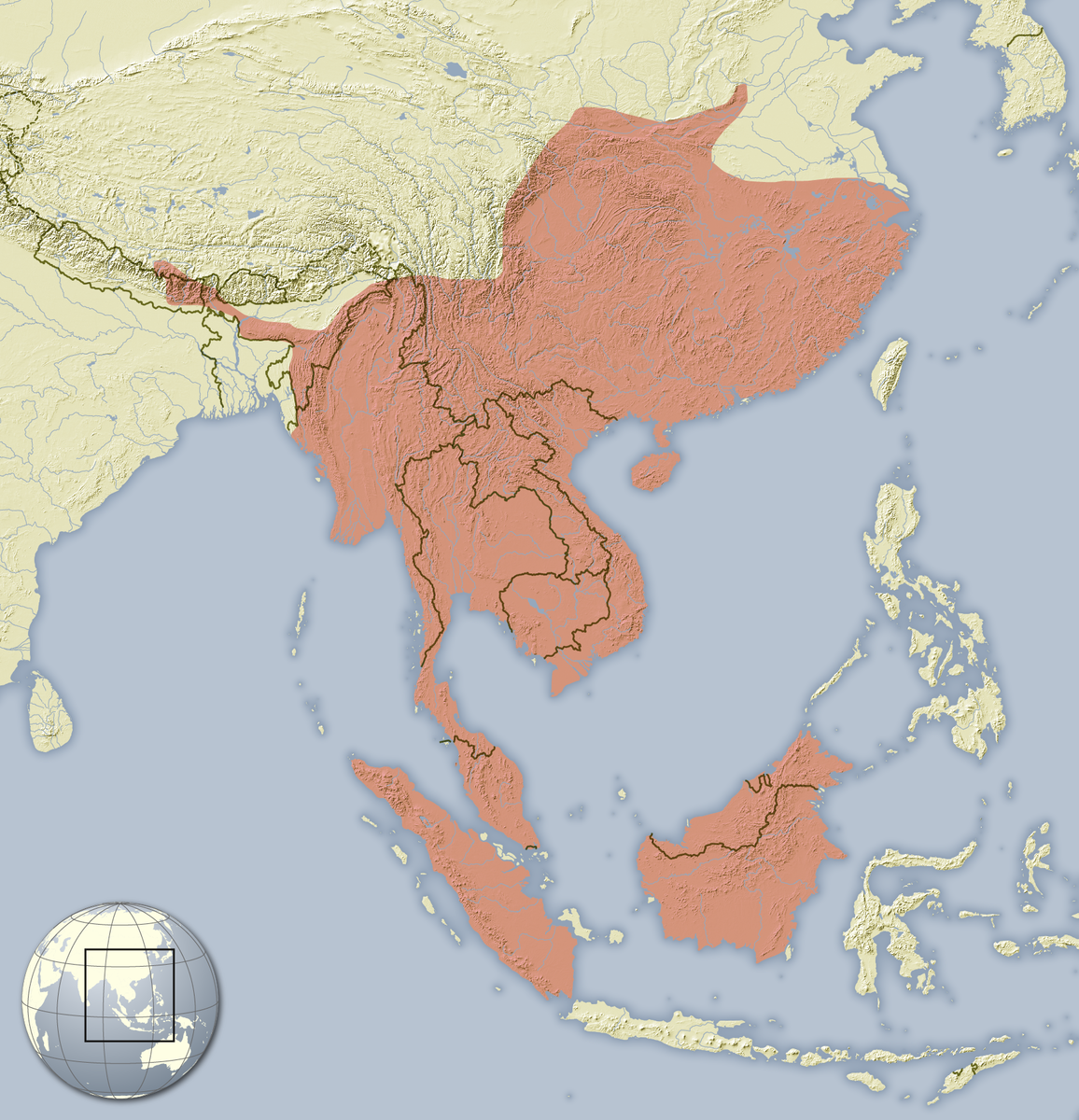
The Hystrix brachyura is distributed throughout Southern Asia, ranging from China, to Thailand, Vietnam, Cambodia, Malaysia, Singapore and Indonesia [1].
Physical Description
This porcupine has short legs, all covered with brown hair. Furthermore, their front legs have four claws, whereas their hind legs have five claws. The Hystrix brachyura is rather short as it measures approximately 67 cm with a tail of 8.5 cm [1].
Reproduction
Regarding reproduction, their gestation period lasts up to 110 days, resulting in a litter size of two to three. Furthermore, anually, the Hystrix brachyura gives birth to two litters [1].
Habitat and Ecology
These terrestrial animals generally live in forest habitats or near argicultural areas. They live in rather small groups, dig out and live in burrows [1].
Behaviour and Diet
Their behaviour is rather remarkable, since they forage at night and sleep during the day. As previously mentioned, they live in small groups, single or in pairs, and can live up to 27 years. Furthemore, they have the ability to swim. Regarding their diet, these porcupines feed on roots, tubers, bark, carrion, insects, large tropical seeds and fallen fruits [1].
About
Selenoproteins
The importance of Selenoproteins
A brief description of selenoproteins
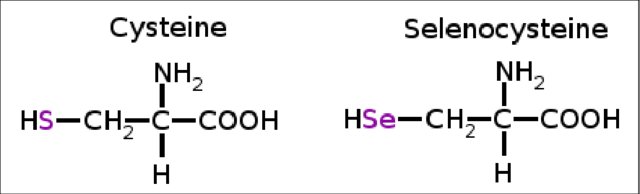
Selenium (Se), the essential trace element, also considered the 21st amino acid, plays an important role in several biological functions such as thyroid hormone metabolism, antioxidant defense systems, the adaptive and acquired immune system and prevention of cancers. Selenoproteins incorporate selenium-containing amino acids. This amino acid is similar to cysteine but instead encloses a selenol group [4]. For its translation, a unique machinery is required. Selenocysteine (Sec) has an analog, cysteine (Cys). Structurally, they are rather similar. However, in Cys' side chain a sulfur (S) atom can be found, while in Sec's side chain, a selenium (Se) can be found.
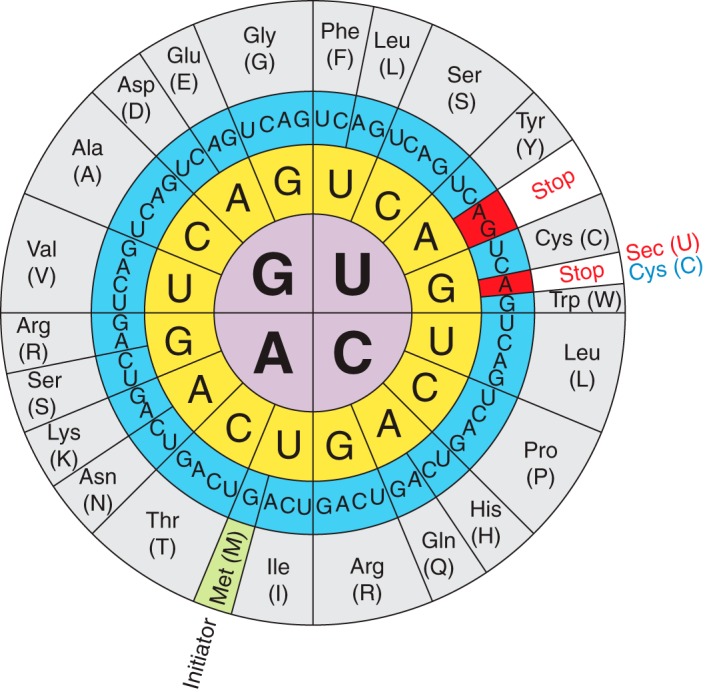
Regarding its translation, a remarkable aspect can be observed: encoded Sec is encoded by the UGA codon, the normally decoded stop codon. However, the precise mechanism of UGA-selenocysteine recoding remains complex, as well as the function of all selenoproteins.
Biosynthesis of Selenoproteins
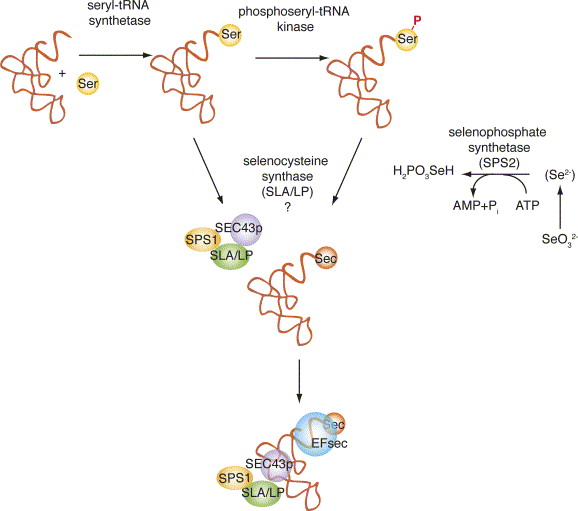
The biosynthesis of selenoproteins is rather complex: The first step in the biosynthesis of the Selenoproteins is the aminoacylation of the tRNA[Ser]Sec with serine thanks to seryl-tRNA synthetase. Seryl moiety is formed and then is converted to phosphoserine by O-phosphoseryl-tRNA kinase (PSTK) and O-phosphoseryl-tRNA[Ser]Sec is formed. Then, SecS then uses selenophosphate as donor of selenium, to create Sec-tRNA[Ser]Sec. Selenophosphate is previously synthesized from selenide and ATP by SPS2 [7].
Families
In this section, the different selenoprotein families will be discussed. Generally, they can also be divided in selenoproteins, cys-homologues and machinery genes. These are the 25 selenoprotein genes: GPx1, GPx2, GPx3, GPx4, GPx6, DI1, DI2, DI3, TR1, TR2, TR3, Sel15, SelH, SelI, SelK, SelM, SelN, SelO, SelP, SelR1, SelS, SelT, SelV, SelW, and SPS2. These are the 10 Cys-homologues: GPx5, GPx7, GPx8, MsrA, SelR2, SelR3, SelU1, SelU2, SelU3 and SelW2. These are the following machinery genes: pstk, SecS, SBP2, secp43, eEFSec, SPS1 and SPS2. However, each protein familiy will now be discussed.
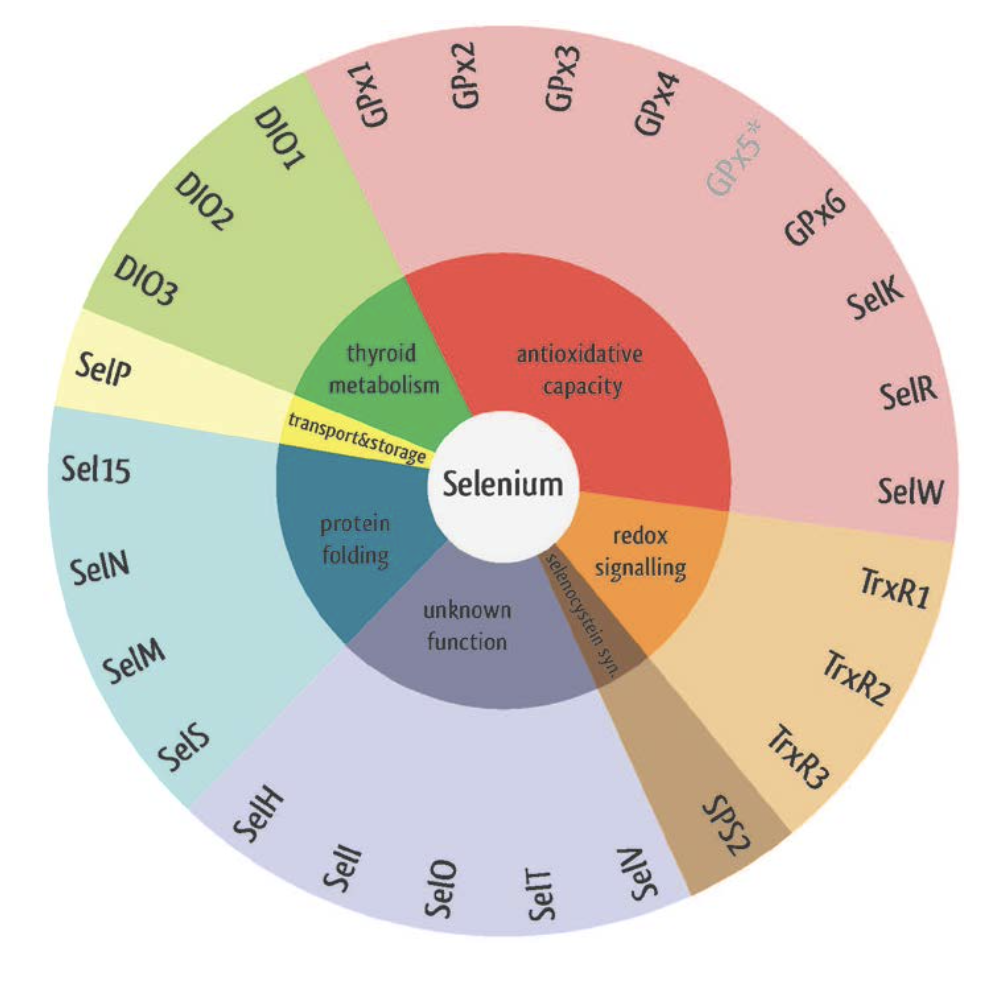
Glutathione peroxidase (GPx)
Gpx family are widespread in all 3 domains of life. They are considered the most important selenoproteins. The GPx family neutralizes reactive oxygen and nitrogen species. They are therefore involved in hydrogen peroxide signaling, detoxification of hydroperoxides and the maintenance of cellular redox homeostasis. There exist 8 different GPx paralogs in mammals: GPx1, GPx2, GPx3, GPx4, GPx5, GPx6, GPx7 and GPx8. [4].
Iodothyronine deiodinase (DIO)
The DIO family consists of 3 different DIO isoforms in mammals: DIO1, DIO2 and DIO3, which are involved in thyroid hormone refulation. They catalyze the release of iodine directly from the thyronine hormones. They furthermore contribute to the (in)activation of the thyroxine and triiodothyronine hormones. DIO1 and DIO2 initiate the thyroid hormone action by converting triiodothyronine to thyroxine whereas DIO3 inactivates triiodothyronine and thyroxine irreversibly [4].
Thioredoxin reductase (TXNRD)
The TXNRD family consists of 3 different TR paralogs in mammals: TR1, TR2 and TR3. Thioredoxin are enzymes known to rgulate redox processes in the cell, such as DNA metabolism and repair and cell signalling. Thus thioredoxin reductases are oxidoreductases that are responsible for disulfide reduction in the cell. The 3 selenoenzymes in this familiy can be characterized regarding its location in the cell [4]:
1) TR1: located in the cytosol
1) TR2: located in the mitochondria
1) TR3: testis-specific
Methionine sulfoxide reducatases A (MsrA)
Methionine sulfoxide reductase A (MSRA) zinc-containing selenoprotein that acts as an antioxidant enzyme that catalyzes the reduction of methionine-S-sulfoxide (MSO) to methionine in proteins and free amino acids [4].
Selenoprotein P (SelP)
Selenoprotein P (SelP) accounts for approximately 50% of the entirety of Se found in plasma. Uniquely, SelP conteins multiple Sec residues and is mainly synthesized in the liver. Together with GPx3, SEL P play an important role in selenium transport and distribution [6].
Selenoprotein R (SelR)
Sel R, also known as methionine-sulfoxide reductase B1 (MsrB1) belongs to the Methionine-R-Sulfoxide Reductase family, which includes three proteins, one of them containing selenocysteine (MSRB1) and two of them being homologues with cysteine (MSRB2 and MSRB23). The MSr family has an antioxidant function that reduces oxidized methionine residues on proteins [4].
Selenoprotein I (SelI)
Selenoprotein I (SelI) is a transmembrane protein encoded by a CDP-alcohol phosphatidyltransferase class-I family gene. It has recently evolved and can be found solely in vertebrates. Furthermore, its function is to produce phosphatidylethanolamine - needed for the formation of vesicular membranes- [6].
Selenoprotein N (SelN)
Selenoprotein N is an ER-resident transmembrane glycoprotein, expressed during the embryonic development and in adult skeletal muscle tissues [6].
Selenoprotein O (SelO)
It contains a single Sec residue. However, the function of SelO remains unknown [6].
Selenoprotein K & S (SelK & SelS)
Sel K and SelS could be in the same family based on their topology, which consists of a transmembrane domain in the NH2-terminal sequence. Both are localized to the ER membrane and have been implicated in ER-associated degradation of misfolded proteins [6].
Seleoprotein U (SelU)
The function and structure of this family remain unknown
SECIS-Binding protein 2 (SBP2)
SBP2 is ubiquitously expressed in all tissues [4].
Selenophosphate synthetase (SPS)
SPS proteins are involved in the synthesis of seleno-phosphate molecules. Thus, they are responsible for the synthesis of Sec-tRNA. Furthermore, they also act as regulators of selenium bioavailability [4].
Selenocysteine synthase (SecS)
SecS protein initiates the incorporation of Sec into selenoproteins by dephosphorylating O-phosphoseryl tRNA[Ser]Sec. Furthermore, it also transfers monoselenophosphate to tRNA [4].
Selenocysteine-specific elongation factor (eEFsec)
eEFsec is a GTP-binding protein. It delivers Sec-tRNAsec to the ribosome [4].
Phosphoseryl t-RNA kinase (PSTK)
In presence of ATP and Magnesium, PSTK acts as kinase that phosphorylates a seryl-tRNAsec complex. This acts as a substrate for SecS [4].
tRNA Sec 1 associated protein 1 (SECp43)
SECp43 protein has 2 ribonucleoprotein-binding domains, that together create an RNA-recognition motif. Furthermore, it is located in the nuclear compartment. Its function contains the regulation of GPx1 and methylated Sec tRNA[Ser]Sec. Lastly, this protein contributes to the formation of the EFSec-SBP2-Sec tRNA[Ser]Sec complex [4].
Methods
The aim of our project is to identify and annotate Hystrix brachyura‘s selenoproteins and other related genes using different bioinformatic tools.
Setup of the computer
The very first step we had to perform in order to set up the computer correctly was to paste the following command in the linux terminal, so all the needed programs and files would be available in the computer:
export PATH=$PATH:/mnt/NFS_UPF/soft/ncbi-blast-2.7.1+/bin/:/mnt/NFS_UPF/soft/exonerate/exonerate-2.2.0-x86_64/bin:/mnt/NFS_UPF/soft/tcoffee/bin:/mnt/NFS_UPF/soft/genewise/x86_64/bin:/mnt/NFS_UPF/soft/fasta
Selection of species
We decided to compare the Hystrix brachyura's genome to the genome of the Cavia Porcellus since it is part of Caviomorpha, which is the closest related family to the Hystricidae family, to which Hystrix brachyura belongs. Another reason why we decided to choose this organism is because its selenoproteins are well characterized in the SelenoDB database. However, there were some proteins that were not found in the Cavia porcellus' genome. This is the reason why we looked for these proteins in the genome of Mus musculus which is another very close relative. Finally we couldn’t find SelW2, thus we took the Homo Sapiens' genomes as the reference genome.
Obtention of Hystrix brachyura genome
We downloaded the Hystrix brachyura's genome and other important files needed to use the different programs from a directory provided by the teachers of the Bioinformatic subject:
/mnt/NFS_UPF/soft/genomes/2021/Hystrix_brachyura
Query acquisition
The sequence of queries has been obtained from SelenoDB 2.0 database. We have downloaded all the 50 query protein sequences into a multifasta file named multifastacavia.fa. It is important to mention that we have downloaded all the proteins contained in the database, these are both the selenoproteins and other related proteins. In this document we changed all the “U” we found for “X” in order to perform a correct analysis, since the software does not recognise it properly. Moreover, all the symbols (such as $ or &) found at the end of the sequences were removed. We did this step manually. The next step was to split the multi fasta file in different single fasta files each containing the sequence of one query protein from the Cavia porcellus. To do so, we used a python script split_multi_fasta.py. In the terminal we have called the next command:
python ~/split_multi_fasta.py -m ~/multifastacavia.fa
As a result we obtained different fasta files named after query.fa, where query is the name of each protein of Cavia porcellus, Mus musculus or Homo sapiens.
tBLASTn
We found out that in order to be able to perform the tblast, we needed to remove some spaces that were present in the name of the query.fa files. To do so we used the following command in the specific directory that contained the file we were interested in removing the spaces:
for f in ./*; do mv "$f" "${f// /_}"; done
We then performed the tBLASTn for all the query.fa files present in a directory. This program compares protein queries against translated nucleotide databases. To execute it, we used the next command:
for file in ./*; do tblastn -query $file -db /mnt/NFS_UPF/soft/genomes/2021/Hystrix_brachyura/genome.fa -outfmt 6 -out ~/step2_blast_outputformat6/$file.blast;done
We want to highlight the fact that we have used the output format 6 where we obtained a table containing different useful values such as the scaffold where the hit is found, the start and length of the alignment in the query and in the reference genome, among other values.
Selection of the regions of interest
From the tblastn we could obtain different pieces of information regarding the significant hits it had found. The best hits are located at the top of the list since they have the highest score value and the smallest E-value. We decided we would be considering as significant the first hit of the list since it is the one with the best values. Not only that, to make it more complete we decided to also get the other hits which were located at the same scaffold as the first one of the list. We did not mind the score of these other hits as long as they were located at the same scaffold. As a result, our region of interest contains all the different hits present in a single scaffold.
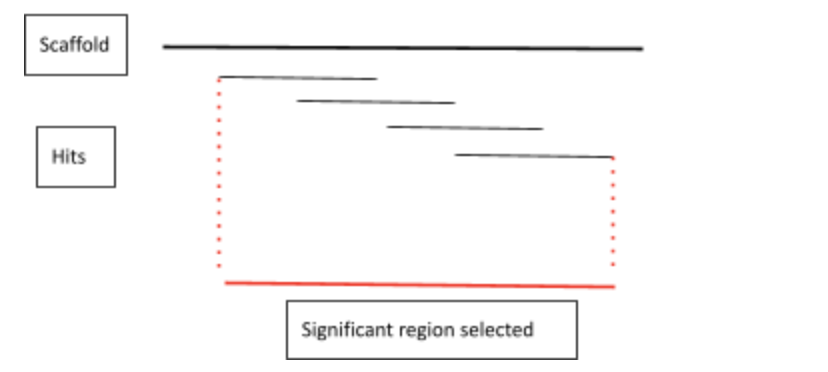
Fasta fetch
Once we had decided the region of interest containing the significant hits we needed to extract the genomic region (scaffold) containing this region. To do so, we used the fastafetch command. This command necessarily needs an indexed genome to work properly. We were given this index in the next directory:
/mnt/NFS_UPF/soft/genomes/2021/Hystrix_brachyura/genome.index
In order to do the fasta fetch we needed some previous steps to be done. First, we created a document containing the name of the scaffold selected for each reference protein. To do so we used the following awk command:
awk '{if(FILENAME!=prev){print $2; close(prev)};prev=FILENAME}' *.blast > tblastn.txt input="./tblastn.txt"
Using this command we accessed a folder containing all the blast files and from each file in it we copied the first cell of the second column (containing the name of the scaffold we decided we wanted to select) and we created a file named tblastn.txt with all the different names of the scaffolds. Once this file was created we executed the following command:
awk '{file=$0 ".fa"; system("fastafetch /mnt/NFS_UPF/soft/genomes/2021/Hystrix_brachyura/genome.fa /mnt/NFS_UPF/soft/genomes/2021/Hystrix_brachyura/genome.index " $0 " > " file.fa)}' tblastn.txt
This allowed us to perform the fastafetch in all of the scaffolds that we had selected as the region of interest. The output files were different fasta files corresponding to each scaffold that contained a significant hit(s). We obtained 40 files, since different query proteins had the hit in the same scaffold.
Fasta subseq
The next step was to obtain a more delimited region where the hits were located. To do so, we used the fasta subseq command.
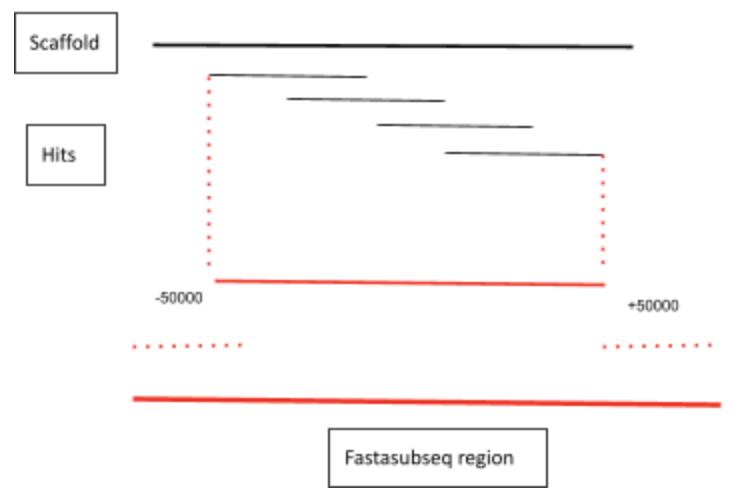
In order to obtain the whole region of interest that contained the gene of the protein, the start and the length of each significant region containing the best hits have been extended 50,000 nucleotides per site.It is important to mention that the start can not be a negative value, that is why we manually set the first nucleotide position to 0. In order to do so, we created a table with important information that could be used in the following steps. To come up with this table we used a Perl script. This table was called coordenades.tbl and it contained the name of the protein, the scaffold and the start and length of the scaffold. It has to be considered that the start and end given are the following:
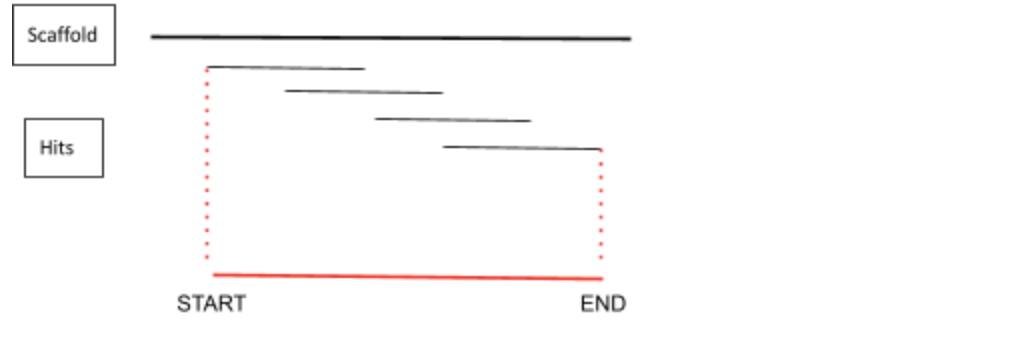
Once we had all this information contained in the table we could execute the following command:
awk '{ print "fastasubseq", $2".fa", $3-50000, ($4-$3)+50000, ">" , $1 "_"$2 "_"$3 ".fa"}' coordenades.tbl > sendfastasub.sh
Then we execute the sh sendfastasub.sh on the terminal. When we executed this command we obtained some particular cases where the last position exceeded the scaffold length. Concretely it was for scaffolds: QZML01083744.1, QZML01141614.1, QZML01139197.1 and QZML01069438.1. To solve this problem we performed the fasta subseq individually setting the last nucleotide position to the last position of the scaffold. We finally obtained all the different fasta files containing the region we had previously decided with 50000 nucleotides less downstream and 50000 more in the length value.
Exonerate
The next step was to obtain just the exonic part of the region of interest, to do so we executed exonerate and then the egrep command to fuse all the exons into the same file.
awk '{ print "exonerate -m p2g --showtargetgff -q", $1 ".fa", "-t" , $1 "_" $2 "_" $3 ".fa" "| egrep -w exon" " >", $1 "_" $2 ".exonerate"".gff"}' coordenades.tbl > sendexonerate.sh
Then we execute the sh sendexonerate.sh on the terminal and we obtained differents files named “exonarate.gff” that contained the information about the exons of the protein of interest.
FastaseqfromGFF
Once we had the files obtained from exonerate we need to get the cDNA sequence of the predicted protein, to do so we use the fastaseqfromGFF command:
awk '{ print "fastaseqfromGFF.pl ", $2".fa", $1".fa", "exonerate.gff", ">", $1 "_" $2 ".nuc"}' coordenades.tbl > sendfastaseqfromGFF.sh
Then we execute the sendfastaseqfromGFF.sh on the terminal and we obtained the different files containing the cDNA sequence of the different proteins of interest.
Fastatranslate
The next step was to translate the cDNA sequence into a protein sequence. The command executed was:
awk '{ print "fastatranslate -f ",$1"_"$2".nuc", "-F 1", " >" $1"_"$2".aa"}' coordenades.tbl > sendfastatranslate.sh
Then we execute the sendfastatranslate.sh on the terminal and we obtained the different files containing the protein sequences.
Substitution of * for X
We needed to change the * corresponding to the selenocysteines predicted by the fastatranslate to X since otherwise the T-Coffee aligns them with a gap in the previous position instead of with the selenocysteine. We changed these symbols manually in the different files.
T-Coffee (Tree-based Consistency Objective Function for alignment Evaluation)
The last step we followed in order to get the alignments was the execution of the T-coffee. In this step we compare the genome of Hystrix Brachyura with the different proteins obtained from Cavia porcellus.
awk '{ print "t_coffee ", $1"_"$2".aa", $1 ".fa", " >", $1"_"$2 "_tcoffee" ".txt"}' coordenades.tbl > sendtcoffee.sh
Then we executed the sendtcoffee.sh on the terminal and we obtained all the alignments.
Genewise
In those cases where the exonerate did not perform well, we proceed to execute the Genewise program. The command used if the orientation of the strand was positive we used the following command:
genewise -pep -pretty -cdna -gff contig_subseq.fa /mnt/NFS_UPF/soft/genomes/2021/Hystrix_brachyura/genome.fa
When the orientation of the strand was negative we used the next command:
genewise -pep -pretty -cdna -gff -trev contig_subseq.fa /mnt/NFS_UPF/soft/genomes/2021/Hystrix_brachyura/genome.fa
SECIS i Seblastian
We used the Seblastian webpage to look for more information about the possible selenoproteins: First, we looked for SECIS elements since they are responsible for the synthesis of the selenocysteine codon from the UGA codon (which is usually a STOP code). If these elements are present in the protein it potentially means that that protein is a selenoprotein. We also used Seblastian to look for prediction of Selenoproteins in other species. It scans upstream of each SECIS for selenoprotein coding sequences with known homologues using blastx, and then it tries to refine the predictions using exonerate.
Results
In the table below you can find the results obtained: all the selenoproteins and seleneprotein's machinery we have found in Hystrix brachyura's genome by aligment to Cavia porcellus' known selenoproteins. An explanation and interpretation can be found in the discussion.
Key:
 Result obtained. Click on the image to consult it.
Result obtained. Click on the image to consult it.  No Result obtained.
No Result obtained.- Protein: Name of the protein
- Species: Name of the specie nearest to Hystrix brachyura used for the alingment
- Residue: If the protein contains Sec (selencystein) or Cys (cystein) in the reference specie
- Scaffold: ID of the scaffold in Hystrix brachyura's genome
- Gene Location: Location of the scaffold within Hystrix brachyura's genome
- Query: Click on the image to download the sequence from C.porcellus used to aligne with Hystrix brachyura's genome
- Orientation: The orientation of the sequence, F for Forward, R for Reverse.
- Tblastn: Results of the comparision of the protein query sequence to a nucleotide sequence - protein to protein comparisons.
- Exonerate: File obtained using exonerate command
- Number of Exons: Number of exons predicted by exonerate
- Protein prediction: The prediction of the Hystrix brachyura's protein sequence
- T-coffee: Alingment between the query from C.porcellus and the protein prediction from Hystrix brachyura
- Seblastian: Results obtained from a selenoprotein prediction server.
- SECIS candidate: SECIS sequences predicted by Seblastian
- Sec: Sec is found both in human and Hystrix brachyura's proteins
- Sec gain: Sec is found in the Hystrix brachyura's protein but not in the human's protein
- Cys: Cystein conteining homolog
- -: There is a gap where a Sec should be
Discussion
Finding an explanation to our results
Initial remarks
Before starting the discussion part we must take into account that in some particular cases we realized that for each subseq we were obtaining more than one protein. We could notice it when we performed the exonerate since we could find exons in two different orientations. So this means that these exonerate files were misleading. We assume that this error is due to the fact that the region of interest chosen was too large. To decide this region we basically took all the different hits that were on the same scaffold (concretely the one that also contained the best hit). To improve it, we could have set a limit in the e-value so the regions we would have obtained as regions of interest would have been smaller and more accurate. In order to try to find some solutions we performed again the exonerate using also the flag -n 1 (exonerate -n 1 -m p2g) so it would give us just the best result. Using this new command we obtained the exonerate file that are the ones that are commented in the Discussion part. However, the data attached in the results part does not contain these new exonerates. We also must say that the alignments that will be commented are obtained with the wrong exonerates. So we expect that if we could do the T-coffee with the new exonerates, the alignements that we would get would be fairly better. It would also be interesting to determine if this second protein that we also had in the fasta seq is a new selenoprotein or if we already detect it somewhere else.
Selenoproteins
DI1
A prediction in the scaffold QZML01000645.1 was selected using the DI1 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50003 and 64225 in the reverse strand. It contains 4 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted DI1 and the Cavia porcellus DI1. It is also relevant the fact that both proteins start with a Metionin (M), this informs us that it is not probable that we have lost information of any part of the protein and that the alignment is very good since all the exons were predicted correctly. The predicted protein has a Sec residue aligned with a Sec residue of the Cavia porcellus DI1, indicating that it is a Selenoprotein. Moreover, we also obtained a good prediction using Seblastian, which used Octodon Degus's as query protein. Regarding the SECIS, a grade A SECIS was found in the positions 4528883-4528950 and a grade B SECIS in positions 56822-56887 was found at the 3’ end both located on the reverse strand. Taken all these results together we can conclude that the gene of DI1 is present in the genome of Hystrix brachyura. We can also say that the predicted protein is a selenoprotein and an orthologous of the Cavia porcellus DI1.
DI2
A prediction in the scaffold QZML01001556.1 was selected using the DI2 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 58325 in the reverse strand. It contains 2 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted DI2 and the Cavia porcellus DI2. It is also relevant the fact that both proteins start with a Metionin (M), this informs us that it is not probable that we have lost information of any part of the protein. The predicted protein has a Sec residue aligned with a Sec residue of the Cavia porcellus DI2. Regarding the SECIS, a grade A SECIS was found in the positions 45004-45078, and a grade B SECIS in positions 24218-24299 was found at the 3’ end both located on the reverse strand. However, we haven't found any prediction with Seblastian. Although we haven't found any good alignment using Seblastian prediction, considering all the other results we can conclude that the gene of DI2 is present in the genome of Hystrix Brachyura. We can also say that the predicted protein is a selenoprotein because we can find a Sec, a SECIS and a good alignment with DI2 from Cavia porcellus so it is an orthologous of the Hystrix brachyura DI2.
DI3
A prediction in the scaffold QZML01001331.1 was selected using the DI3 Mus musculus protein as a query based on our criteria. We used this species since we could not find this protein in the genome of Cavia porcellus. The predicted gene is located between the positions 645907 to 646731 in the reverse strand. It contains 1 exon. The T-coffee made a good alignment, the score was high (1000) between the predicted DI3 and the Mus musculus DI3. It is also relevant the fact that both proteins start with a Metionin (M), this informs us that it is not probable that we have lost information of any part of the protein so we are comparing all of the exons. The predicted protein has a Sec residue aligned with a Sec residue of the Mus musculus DI3. We have found a total of 7 SECIS. There is a SECIS grade A located in positions 45272 to 645352in the reverse strand. Then we can also find 5 grade B SECIS and 1 grade C SECIS. Using Seblastian we also obtained a good prediction using Chinchilla lanigera’s genome as query protein. This result strongly suggests that the DI3 gene is conserved in the Hystrix brachyura genome and it is still a selenoprotein. We can say that DI3 from Mus musculus is an orthologous version of the Hystrix brachyura DI3.
GPx1 (not found in the genome)
A prediction in the scaffold QZML01000118.1 was selected using the DI3 Mus musculus protein as a query based on our criteria. However, this protein (GPx1) is only found in Mus Musculus and not in the Cavia porcellus and thus, it is not likely that Hystrix brachyura has it since, in terms of evolution, it is closer to Cavia porcellus than to Mus Musculus. Our results show that this protein can be discarded since Exonerate do not predict the protein in this scaffold. Then, we can conclude that the predicted GPx1 is not found in the Hystrix brachyura genome.
GPx2
A prediction in the scaffold QZML01000308.1 was selected using the GPx2 Mus musculus protein as a query based on our criteria. We used this species since we could not find this protein in the genome of Cavia porcellus. The predicted gene is located between the positions 50000 and 52818 in the forward strand. It contains 2 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted GPx2 and the Mus musculus GPx2. It is also relevant the fact that both proteins start with a Metionin (M), this informs us that it is not probable that we have lost information of any part of the protein so we are comparing all of the exons. The predicted protein has a Sec residue aligned with a Sec residue of the Mus musculus GPx2. However, no SECIS element was found. Taking all this information into account we can say that the gene of GPx2 is conserved in H. brachyura genome. However we can not assure that it is still a selenoprotein because although it contains a Sec residue the SECIS element is missing. So potentially can have happened two things:
1) Maybe the SECIS element exists but it has not been predicted correctly, so it is a selenoprotein.
2) Or the SECIS does not exist so is not a selenoprotein since the machinery is not able to be recruited.
So we can not be sure if GPx2 is still a selenoprotein.
GPx3
A prediction in the scaffold QZML01000499.1 was selected using the GPx3 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 52653 in the reverse strand. It contains 4 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted GPx3 and the Cavia porcellus GPx3. It is also relevant the fact the alignment does not start with a Metionin (M). However, both proteins start with a Q aminoacid, so we do not expect that any information has been lost. The predicted protein has a Sec residue aligned with a Sec residue of the Cavia porcellus GPx3. Moreover, we obtained a good prediction with Seblastian, which used Dipodomys ordii genome to compare. Regarding the SECIS, a grade A SECIS was found in the positions 49381-49458. It was found at the 3’ end of the negative strand. Taken all these results together we can conclude that the gene of GPx3 is present in the genome of Hystrix brachyura. We can also say that the predicted protein is a selenoprotein and an orthologous of the Cavia porcellus GPx3.
GPx4
A prediction in the scaffold QZML01000690.1 was selected using the GPx4 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 50358 in the reverse strand. It contains 2 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted GPx4 and the Cavia porcellus GPx4. It is also relevant the fact that both proteins do not start with a Metionin (M). However, both proteins start with a V aminoacid, so we do not expect that any information has been lost. The predicted protein has a Sec residue aligned with a Sec residue of the Cavia porcellus GPx4. Moreover, we obtained a good prediction with Seblastian, which used Octodon degus's genome to compare. Regarding the SECIS, a grade A SECIS was found in the positions 49463-49541 was found at the 3’ end of the negative strand. Taken all these results together we can conclude that the gene of GPx4 is conserved in the genome of Hystrix brachyura. We can also say that the predicted protein is a selenoprotein and an orthologous of the Cavia porcellus GPx4.
GPx5
A prediction in the scaffold QZML01003264.1 was selected using the GPx5 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 53251 in the reverse strand. It contains 4 exons. We first realized a T-coffee alignment using Exonerate. Although the score was high (999) we obtained a large fragment of the initial part of the protein that was not aligned properly. We also could see that H. brachyura has some Sec residues but were not aligned properly with the reference protein of Cavia porcellus. In order to try to improve the alignment we performed a Genewise. The T-coffee of Genewise made a better alignment. Although the score was lower (992) we could now align the Sec residue from H. brachyura. It is very important to notice that this Sec residue is aligned with a Cys residue in the protein from Cavia porcellus. This means that a Sec gain has occurred in H. brachyura. We can also see that this protein was a Cys-homologous protein in Cavia porcellus. However, we do not obtain any prediction with Seblastian and any SECIS. Taking all this information into account we can say that the gene of GPx5 is conserved in H. Brachyura genome. However we can not assure that it is still a selenoprotein because although it contains a Sec residue the SECIS element is missing. So potentially can have happened two things:
1) Maybe the SECIS element exists but it has not been predicted correctly, so it is a selenoprotein.
2) Or the SECIS does not exist so is not a selenoprotein since the machinery is not able to be recruited.
So we can not be sure if GPx5 is still a selenoprotein.
GPx6
A prediction in the scaffold QZML01003264.1 was selected using the GPx6 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 70504 and 73955 in the forward strand. It contains 4 exons. We first realized a T-coffee alignment using Exonerate. Although the score was high (999) we obtained a large fragment of the initial part of the protein that was not aligned properly. We also could see that H. brachyura has some Sec residues but were not aligned properly with the reference protein of Cavia porcellus. In order to try to improve the alignment we performed a Genewise. The T-coffee of Genewise made a better alignment. Although the score was lower (997) we could now align the Sec residue from H. Brachyura. It is very important to notice that this Sec residue is aligned with a Cys residue in the protein from Cavia porcellus. This means that a Sec gain has occurred in H. brachyura. We can also see that this protein was a Cys-homologous protein in Cavia porcellus. However, we do not obtain any prediction with Seblastian and any SECIS. Taking all this information into account we can say that the gene of GPx5 is conserved in H. Brachyura genome. However we can not assure that it is still a selenoprotein because although it contains a Sec residue the SECIS element is missing. So potentially can have happened two things:
1) Maybe the SECIS element exists but it has not been predicted correctly, so it is a selenoprotein.
2) Or the SECIS does not exist so is not a selenoprotein since the machinery is not able to be recruited.
So we can not be sure if GPx6 is still a selenoprotein.
GPx7
A prediction in the scaffold QZML01000458.1 was selected using the GPx7 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 51949 in the forward strand. It contains 2 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted GPx7 and the Cavia porcellus GPx7. It is also relevant the fact that both proteins do not start with a Metionin (M). However, both proteins start with a Q aminoacid, so we do not expect that any information has been lost. Neither the predicted protein nor the protein of the Cavia porcellus have any Sec residue. However they contain Cys residues. Moreover, we do not obtain any prediction with Seblastian and any SECIS. Taken all these results together we can conclude that the gene GPx7 of Cavia porcellus is present in his genoma, but it is not a selenoprotein but a Cys-containing homologue (just the same as the GPx7 protein in Cavia porcellus).
GPx8
A prediction in the scaffold QZML01000632.1 was selected using the GPx8 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 52460 in the forward strand. It contains 2 exons. The T-coffee made a good alignment, the score was high (999) between the predicted GPx8 and the Cavia porcellus GPx8. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with a different aminoacid, so maybe we have lost part of the protein. Neither the predicted protein nor the protein of the Cavia porcellus have any Sec residue. However they contain Cys residues. Moreover, we do not obtain a prediction with Seblastian. However a grade B SECIS was found in the positions 6359-6432. It was found at the 3’ end of the reverse strand. Taken all these results together we can conclude that the gene GPx8 of Cavia porcellus is present in his genoma, but it is not a selenoprotein but a Cys-containing homologue (just the same as the GPx8 protein in Cavia porcellus). Nevertheless, we must take into account that a SECIS is found, however is a grade B SECIS so our hypothesis is that maybe the prediction of SECIS is not correct.
MsrA
A prediction in the scaffold QZML01083744.1 was selected using the MsrA Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 161 and 292 in the forward strand. It contains 1 exon. The T-coffee made an alignment with a score of (980) between the predicted MsrA and the Cavia porcellus MsrA. If we take a look at the t-coffee obtained we can see the alignment is not very good. We guess MsrA was a cys-homologue in Cavia porcellus and it continues being it in the H. brachyura genome. Due to the fact that the alignment is not good enough we can not be sure. However, we can not find any Sec residue and neither SECIS nor Seblastian so we can conclude this is not a Selenoportein.
Sel15
A prediction in the scaffold QZML01001257.1 was selected using the Sel15 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 60578 and 91435 in the forward strand. It contains 4 exons. The T-coffee made a good alignment, the score was high (999) between the predicted Sel15 and the Cavia porcellus Sel15. It is also relevant the fact that both proteins do not start with a Metionin (M). They start with a V aminoacid and we can see that the first part of the alignment is not perfect since there is a big gap so we could use Genewise in order to get this part better. The predicted protein has a Sec residue aligned with a Sec residue of the Mus musculus Sel15. However, no SECIS element was found nor was Seblastian prediction. Taking all this information into account we can say that the gene of Sel15 is conserved in H. Brachyura genome. However we can not assure that it is still a selenoprotein because although it contains a Sec residue the SECIS element is missing. So potentially can have happened two things:
1) Maybe the SECIS element exists but it has not been predicted correctly, so it is a selenoprotein.
2) Or the SECIS does not exist so is not a selenoprotein since the machinery is not able to be recruited.
So we can not be sure if Sel15 is still a selenoprotein.
SelK
A prediction in the scaffold QZML01000202.1 was selected using the SelK Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 52145 in the forward strand. It contains 3 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted SelK and the Cavia porcellus SelK. It is also relevant the fact the alignment does not start with a Metionin (M). However, both proteins start with a G aminoacid, so we do not expect that any information has been lost. The predicted protein with exonerate has a GAP residue aligned with a X residue of the Cavia porcellus SelK. This is because the alignment has not been correctly performed, since the last part of the sequence of the Hystrix brachyura is a GAP. This indicates that the alignment is not correct and the genewise should be done so we could potentially obtain a better alignment where the Sec residu aligns with a residue of the Hystrix brachyura sequences. In conclusion, we do not have enough information. Moreover, we do not obtain a prediction with Seblastian. However a grade B SECIS was found in the positions 51491-51558. It was found at the 3’ end of the forward strand. Taken all these results together we can conclude that the gene GPx8 of Cavia porcellus is present in his genoma, but it is not a selenoprotein but a Cys-containing homologue (just the same as the GPx8 protein in Cavia porcellus). Nevertheless, we must take into account that a SECIS is found, however is a grade B SECIS so our hypothesis is that maybe the prediction of SECIS is not correct.
SelH
A prediction in the scaffold QZML01001004.1 was selected using the SelH Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 50582 in the forward strand. It contains 3 exons. The T-coffee made a good alignment, the score was high (999) between the predicted SelH and the Cavia porcellus SelH. It is also relevant the fact that both proteins start with a Metionin (M), this informs us that it is not probable that we have lost information of any part of the protein so we are comparing all of the exons. However, we do not obtain any prediction with Seblastian and any SECIS. The predicted protein has a Sec residue aligned with a Sec residue of the Cavia porcellus SelH. Taking all this information into account we can say that the gene of SelH is conserved in H. brachyura genome. However we can not assure that it is still a selenoprotein because although it contains a Sec residue the SECIS element is missing. So potentially can have happened two things:
1) Maybe the SECIS element exists but it has not been predicted correctly, so it is a selenoprotein.
2) Or the SECIS does not exist so is not a selenoprotein since the machinery is not able to be recruited.
So we can not be sure if selH is still a selenoprotein.
SelI
A prediction in the scaffold QZML01141614.1 was selected using the SelI cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 726 and 1001 in the forward strand. It contain 1 exon. The T-coffee made a good alignment, the score was high (993) between the predicted GPx8 and the Cavia porcellus GPx8. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with a different aminoacid, so maybe we have lost part of the protein. In Cavia porcellus we found a Sec residue but we do not found any alignment with the predicted protein. This indicates that the alignment with exonerate is not optimal. However they contain Cys residues. However, we do not obtain any prediction with Seblastian and any SECIS. Taken all these results together we can conclude that the gene SelI of Cavia porcellus is present in his genoma, but it is not a selenoprotein but a Cys-containing homologue (just the same as the SelI protein in Cavia porcellus). Nevertheless, we must take into account that a alignment with exonerate is not totally correct, maybe a genewise.
SelM
A prediction in the scaffold QZML01000350.1 was selected using the SelM Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50024 and 52127 in the reverse strand. It contains 5 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted SelM and the Cavia porcellus SelM. It is also relevant the fact that both proteins do not start with a Metionin (M). However, both proteins start with a Y aminoacid, so we do not expect that any information has been lost. The predicted protein has a Sec residue aligned with a Sec residue of the Cavia porcellus SelM. Moreover, we obtained a good prediction with Seblastian, which used the Dasypus novemcinctus genome to compare. Regarding the SECIS, a grade A SECIS was found in the positions 49884-49956 was found at the 3’ end of the reverse strand. Taken all these results together we can conclude that the gene of SelM is conserved in the genome of Hystrix brachyura. We can also say that the predicted protein is a selenoprotein and an orthologous of the Cavia porcellus SelM.
SelN
A prediction in the scaffold QZML01000262.1 was selected using the SelN Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 62010 in the forward strand. It contains 11 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted SelN and the Cavia porcellus SelM. It is also relevant the fact that both proteins do not start with a Metionin (M). However, both proteins start with a Q aminoacid, so we do not expect that any information has been lost. The predicted protein has a Sec residue aligned with a Sec residue of the Cavia porcellus SelN. However, we do not obtain any prediction with Seblastian and any SECIS. Taking all this information into account we can say that the gene of SelN is conserved in H. brachyura genome. However we can not assure that it is still a selenoprotein because although it contains a Sec residue the SECIS element is missing. So potentially can have happened two things:
1) Maybe the SECIS element exists but it has not been predicted correctly, so it is a selenoprotein.
2) Or the SECIS does not exist so is not a selenoprotein since the machinery is not able to be recruited.
So we can not be sure if SelN is still a selenoprotein.
SelO
A prediction in the scaffold QZML01000349.1 was selected using the SelO Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 58154 in the reverse strand. It contains 9 exons. The T-coffee made a good alignment, the score was high (996) between the predicted SelO and the Cavia porcellus SelO. It is also relevant the fact that both proteins start with a Metionin (M), this informs us that it is not probable that we have lost information of any part of the protein so we are comparing all of the exons. The predicted protein has a Sec residue aligned with a Sec residue of the Cavia porcellus SelO. Moreover, we obtained a good prediction with Seblastian, which used the Chinchilla lanigera genome to compare. Regarding the SECIS, a grade A SECIS was found in the positions 49853-49934 in reverse strand, and a grade B SECIS in positions 21731-21805 in forward strand was found at the 3’ end. Taken all these results together we can conclude that the gene of SelO is conserved in the genome of Hystrix brachyura. We can also say that the predicted protein is a selenoprotein and an orthologous of the Cavia porcellus SelO.
SelP
A prediction in the scaffold QZML01000180.1 was selected using the SelP Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 57328 in the forward strand. It contains 4 exons. In T-coffee with a score (970) we can see many Sec residues in both the sequence of Cavia porcellus and H. brachyura, just a few of them are correctly aligned. We assume that the alignment is not correct and this is the reason why not all the Sec residu are correctly aligned. Maybe the alignment obtained with the new exonerate file would be more precise. We concluded that although some Sec residues can be found in the H. brachyura genome we can not find SECIS nor seblastian prediction. o potentially can have happened two things:
1) Maybe the SECIS element exists but it has not been predicted correctly, so it is a selenoprotein.
2) Or the SECIS does not exist so is not a selenoprotein since the machinery is not able to be recruited.
SelR1
A prediction in the scaffold QZML01001025.1 was selected using the SelR1 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50326 and 50652 in the reverse strand. It contain 1 exon. In T-coffee with a higher score (1000) we can see one Sec residue in both the sequence of Cavia porcellus and H. brachyura are correctly aligned. Also we can see that Hystrix Brachyura has gain a Sec residue because it is aligned with a R aminoacid of the Cavia porcellus genome. Regarding the SECIS, a grade A SECIS was found in the positions 49518-49589 in reverse strand was found at the 3’ end. However, we do not obtain any prediction with Seblastian. Taken all these results together we can conclude that the gene of SelR1 is conserved in the genome of Hystrix brachyura. We can also say that the predicted protein is a selenoprotein and an orthologous of the Cavia porcellus SelR1.
SelR2
A prediction in the scaffold QZML01139197.1 was selected using the SelR2 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 572 and 769 in the reverse strand. It contain 1 exon. The T-coffee with the genewise made a good alignment, the score was high (984) between the predicted SelR2 and the Cavia porcellus SelR2. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with a different aminoacid, so maybe we have lost part of the protein. Neither the predicted protein nor the protein of the Cavia porcellus have any Sec residue. However they contain Cys residues. Moreover, we do not obtain any prediction with Seblastian and any SECIS. Taken all these results together we can conclude that the gene SelR2 of Cavia porcellus is present in his genoma, but it is not a selenoprotein but a Cys-containing homologue (just the same as the SelR2 protein in Cavia porcellus).
SelR3
A prediction in the scaffold QZML01000546.1 was selected using the SelR3 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 173555 in the reverse strand. It contains 4 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted SelR3 and the Cavia porcellus SelR3. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with a G aminoacid, so maybe we have lost part of the protein. Neither the predicted protein nor the protein of the Cavia porcellus have any Sec residue. However they contain Cys residues. Regarding the SECIS, a grade C SECIS was found in the positions 13400-13483 in the forward strand it was found at the 3’ end. We do not consider this SECIS because grade C is not good. Moreover, we do not obtain any prediction with Seblastian. Taken all these results together we can conclude that the gene SelR3 of Cavia porcellus is present in his genoma, but it is not a selenoprotein but a Cys-containing homologue (just the same as the SelR3 protein in Cavia porcellus).
SelS
A prediction in the scaffold QZML01002059.1 was selected using the SelS Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 57907 in the forward strand. It contains 5 exons. The T-coffee made a good alignment, the score was high (999) between the predicted SelS and the Cavia porcellus SelS. Moreover, both proteins start with a M aminoacid, so we do not expect that any information has been lost. However, the predicted protein with exonerate has a GAP residue aligned with a Sec residue of the Cavia porcellus SelS. This is because the alignment has not been correctly performed, since the last part of the sequence of the Hystrix brachyura is a GAP. This indicates that the alignment is not correct and the genewise should be done so we could potentially obtain a better alignment where the Sec residu aligns with a residue of the Hystrix brachyura sequences. In conclusion, we do not have enough information. Moreover, we do not obtain SECIS and also we do not obtain a prediction with Seblastian. Taking all the results into account, we can not conclude that SelS is a selenoprotein but we can tell that the SelS gene is conserved in the H. brachyura genome.
SelT
A prediction in the scaffold QZML01000126.1 was selected using the SelT Cavia porcellus' protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 65493 in the forward strand. It contains 5 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted SelT and the Cavia porcellus SelT. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with a L aminoacid, so maybe we have lost part of the protein. The predicted protein has a Sec residue aligned with a Sec residue of the Cavia porcellus SelT. Regarding the SECIS, a grade B SECIS was found in the positions 44768-44843 was found at the 3’ end located on the forward strand. However, we haven't found any prediction with Seblastian. Although we haven't found any alignment using Seblastian prediction, considering all the other results we can conclude that the gene of SelT is present in the genome of Hystrix brachyura. We can also say that the predicted protein is a selenoprotein because we can find a Sec, a SECIS and a good alignment with SelT from Cavia porcellus so it is an orthologous of the Hystrix brachyura SelT.
SelU1
A prediction in the scaffold QZML01001705.1 was selected using the SelU1 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 7133 and 7828 in the forward strand. It contain 1 exon. The T-coffee made a good alignment, the score was (986) between the predicted SelU1 and the Cavia porcellus SelU1. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with a different aminoacid, so maybe we have lost part of the protein. The predicted protein has a Sec residue aligned with a W residue of the Cavia porcellus SelU1. Regarding the SECIS, a grade B SECIS was found in the positions 48321-48392 was found at the 3’ end located on the reverse strand. However, we haven't found any prediction with Seblastian. Taken all these results together we can conclude that the gene SelU1 of Cavia porcellus is present in his genoma. The results indicate that a Sec gain has occurred in Hystrix brachyura and that it has been converted to a selenoprotein.
SelU2
A prediction in the scaffold QZML01001832.1 was selected using the SelU2 Mus musculus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 68936 in the forward strand. It contains 5 exons. The T-coffee made a good alignment, the score was high (990) between the predicted SelU3 and the Mus musculus SelU3. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with (A) aminoacid, so maybe we have lost part of the protein. Neither the predicted protein nor the protein of the Mus musculus have any Sec residue. However they contain Cys residues. Moreover, we do not obtain SECIS and also we do not obtain a prediction with Seblastian. Taken all these results together we can conclude that the gene SelU2 of Mus musculus is present in his genoma, but it is not a selenoprotein but a Cys-containing homologue (just the same as the SelU2 protein in Mus musculus).
SelU3
A prediction in the scaffold QZML01001819.1 was selected using the SelU3 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 51853 in the reverse strand. It contains 6 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted SelU3 and the Cavia porcellus SelU3. It is also relevant the fact that both proteins do start with a Metionin (M), this informs us that it is not probable that we have lost information of any part of the protein and that the alignment is very good since all the exons were predicted correctly. Neither the predicted protein nor the protein of the Cavia porcellus have any Sec residue. However they contain Cys residues. Moreover, we do not obtain any prediction with Seblastian and any SECIS. Taken all these results together we can conclude that the gene SelU3 of Cavia porcellus is present in his genoma, but it is not a selenoprotein but a Cys-containing homologue (just the same as the SelU3 protein in Cavia porcellus).
SelW
A prediction in the scaffold QZML01000939.1 was selected using the SelW Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 50254 in the forward strand. It contains 2 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted SelW and the Cavia porcellus SelW. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with a G aminoacid, so maybe we have lost part of the protein. Neither the predicted protein nor the protein of the Cavia porcellus have any Sec residue. However they contain Cys residues. Moreover, we do not obtain any prediction with Seblastian and any SECIS. Taken all these results together we can conclude that the gene SelW of Cavia porcellus is present in his genoma, but it is not a selenoprotein but a Cys-containing homologue (just the same as the SelW protein in Cavia porcellus).
SelW2
A prediction in the scaffold QZML01000939.1 was selected using the SelW2 Homo sapiens protein as a query based on our criteria. The predicted gene is located between the positions 49307 and 50320 in the forward strand. It contains 4 exons. The T-coffee made a good alignment, the score was high (1000) between the predicted SelW2 and the Homo sapiens SelW2. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with a G aminoacid, so maybe we have lost part of the protein. Neither the predicted protein nor the protein of the Homo sapiens have any Sec residue. However they contain Cys residues. Moreover, we do not obtain any prediction with Seblastian and any SECIS. Taken all these results together we can conclude that the gene SelW2 of Homo sapiens is present in his genoma, but it is not a selenoprotein but a Cys-containing homologue (just the same as the SelW2 protein in Homo sapiens).
TR1
A prediction in the scaffold QZML01001178.1 was selected using the TR1 Mus musculus as a query based on our criteria. The predicted gene is located between the positions 50104 and 51494 in the forward strand. It contains 1 exon. In T-coffee we can see a very bad alignment with a score of 817 and very tiny parts aligned. We assume it is due to the fact that to do the alignment we have not been able to do it using the corrected exonerate file so the whole alignment is not correct. The predicted protein has many Sec residues aligned with a different residue of the Mus musculus SelU1. Moreover, we did not obtain any prediction from Seblastian. On SECIS we found 6 of grade B, but none relevant for to be considered. Taken all these results together we can not conclude anything from this alignment since we do not think it is correct.
TR2
A prediction in the scaffold QZML01001178.1 was selected using the TR2 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 51408 in the forward strand. It contain 1 exon. The T-coffee made a bad alignment, the score was (827) between the predicted TR2 and the Cavia porcellus TR2. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with a E aminoacid, so maybe we have lost part of the protein. If we take a look at the t-coffee obtained we can see the alignment is not very good. We guess TR2 was a cys-homologue in Cavia porcellus and it continues being in the Hystrix brachyura genome. Due to the fact that the alignment is not good enough we can not be sure. However, we can not find any Sec residue and neither SECIS nor Seblastian so we can conclude this is not a Selenoportein.
TR3
A prediction in the scaffold QZML01001178.1 was selected using the TR3 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 49938 and 51408 in the forward strand. It contain 1 exon. The T-coffee made a bad alignment, the score was high (933) between the predicted TR3 and the Cavia porcellus TR3. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with a different aminoacid, so maybe we have lost part of the protein. We assume it is due to the fact that to do the alignment we have not been able to do it using the corrected exonerate file so the whole alignment is not correct. Moreover, we do not obtain any prediction with Seblastian and any SECIS. However, we should also notice that in the alignment we can see that there are many Sec residues in the H. brachyura genome. One possible explanation of these new Sec residues is that this protein has been gain. In conclusion, we do not have enough information to conclude anything.
Selenoprotein machinery
SBP2
A prediction in the scaffold QZML01000351.1 was selected using the SBP2 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 67521 in the reverse strand. It contains 9 exons. The T-coffee made a good alignment, the score was high (996) between the predicted SBP2 and the Cavia porcellus SBP2. It is also relevant the fact that both proteins do not start with a Metionin (M). Both proteins start with a L aminoacid, so maybe we have lost part of the protein. Neither in the predicted protein nor that of Cavia procellus were Sec residues found. Moreover, we do not obtain any prediction with Seblastian and any SECIS. As we have found no SECIS or Sec residues and the alignment is good, we can conclude that SBP2 is a gene that is part of the selenoprotein machinery.
SPS
A prediction in the scaffold QZML01000677.1 was selected using the SPS Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 49952 and 51211 in the reverse strand. It contains 1 exon. Using T-coffee a good alignment was predicte (Score:998). We can see that both H. brachyura and Cavia porcellus contain a Sec residue. Moreover Seblastian made a selenoprotein prediction using Odobenus rosmarus divergens genome. Not only that, but a SECIS was found in the reverse strand between the positions: 49308-49384. We can conclude that in Cavia porcellus SPS stopped being a protein from selenoprotein machinery and started being a selenoprotein itself. The same happened in Hystrix Brachyura, which makes sense since its close phylogenetic relation.
SecS
A prediction in the scaffold QZML01001130.1 was selected using the SecS Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 82892 in the reverse strand. It contains 11 exons. Using T-coffee a good alignment was made (Score:1000). We can see that a Sec gain has occurred in the H. brachyura genome. However, no Seblastian prediction has been found. We have not obtained any SECIS element prediction. Here we can not conclude that SecS has become a selenoprotein although we have found a Sec residue since no SECIS element is found. This makes us conclude that SecS is still part of the Seleneoprotein machinery.
eEFsec
A prediction in the scaffold QZML01001046.1 was selected using the eEFsec Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 49964 and 244641 in the forward strand. It contains 7 exons. In T-coffee we can see a very bad alignment with a score of 786 and very tiny parts aligned. We assume it is due to the fact that to do the alignment we have not been able to do it using the corrected exonerate file so the whole alignment is not correct. However, we should also notice that in the alignment we can see that there are many Sec residues in the H. brachyura genome. One possible explanation of these new Sec residues is that this protein which is part of the selenoprotein machinery has become a selenoprotein itself. Nevertheless we can not confirm this hypothesis. Moreover, neither SECIS nor Seblastian were found. Taken all these results together we can not conclude anything from this alignment since we do not think it is correct.
PSTK
A prediction in the scaffold QZML01000320.1 was selected using the PSTK Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 54534 in the reverse strand. It contains 4 exons. A good alignment was made (Score: 999) and no Sec residues neither Seblastian prediction were found. Unexpectedly we found a SECIS element located in the forward strand in the positions 27606-27673. We can conclude that this SECIS prediction is not relevant since it is located in the forward strand. So the PSTK gene is conserved in the H. brachyura genome and it is part of the selenoprotein machinery.
SECp43
A prediction in the scaffold QZML01000905.1 was selected using the SECp43 Cavia porcellus protein as a query based on our criteria. The predicted gene is located between the positions 50000 and 50669 in the forward strand. It contains 2 exons. In T-coffee we can see a very bad alignment with a score of 807 and very tiny parts aligned. We assume it is due to the fact that to do the alignment we have not been able to do it using the corrected exonerate file so the whole alignment is not correct. However, we should also notice that in the alignment we can see that there are many Sec residues in the H. brachyura genome. One possible explanation of these new Sec residues is that this protein which is part of the selenoprotein machinery has become a selenoprotein itself. Nevertheless we can not confirm this hypothesis. Moreover, neither SECIS nor Seblastian were found. Taken all these results together we can not conclude anything from this alignment since we do not think it is correct.
Conclusion
The aim of this study is to look for the Selenoproteins and the Selenoprotein machinery that can be found in the Hystrix brachyura’s genome. We have used several bioinformatic tools to compare Hystrix brachyura’s genome with the already described selenoproteins of Cavia porcellus due to phylogenetic proximity. However, in some particular cases we have used Mus musculus’ and Homo sapiens’ genomes to look for some concrete Selenoprotein. The alignment of the sequences of these species with the Hystrix brachyura’s sequence has allowed us to infer some information about the Selenoprotein that this last specie contain. The first result we can deduce from our results is that GPx1 selenoprotein is lost in Hystrix brachyura. Moreover our results allow us to do a classification in different groups of Selenoproteins: The first group is formed by those Selenoprotein genes that are conserved in the Hystrix brachyura’s genome and they still are Selenoproteins since they have the Sec residue and some SECIS elements. Here we can include the next selenoproteins: DI1,DI2,GPx3,GPx4,SelM, SelO,SelR1, SelT. Another group includes those cases where a Sec gain has occurred in the Hystrix brachyura’s genome and we can also find a SECIS element, in these cases we consider these proteins as actual Selenoproteins. This is the case of SelU1. One more group is formed by those Selenoproteins that contain a Sec residue but do not have a SECIS element. Therefore, we can not consider them as actual Selenoproteins. In this group we can include: GPx2, SelH, SelN, SelP, Sel15. We can also define a subgroup where a Sec gain has occurred in Hystrix brachyura’s genome but no SECIS element can be found, so we can not consider them as proper Selenoproteins. This is the particular case of GPx5, GPx6. Then we also have the group of the Cys-containing homologs, here we can include: GPx7,GPx8, SelK,SelR2, SelU2, SelU3,SelW, SelW2 and SelR3. We have also found a group formed by the selenoprotein machinery: PSTK, SecS and SBP2. We have also found a particular case (SPS) where a protein from selenoprotein machinery in other species is indeed a selenoprotein in both Cavia porcellus and Hystrix brachyura. Finally, we have some cases where the information obtained is not enough, so we can not conclude anything about them since more evidence is needed. Here we have the next proteins: SelK, Seli, SelS, MsrA,TR2,TR3, SECp43, eEFsec,TR1.
References
Team
Who are we?