The following thing we did was classifying this results into different categories (using woundedknee.pl) according with possibilities of BLAST matching (we used some parameters to filter the hits that are not significant):
- Two exons: in this case our query will match with two separated regions of the same subject (about 50 pb with one region and 50 pb with another one). So, there is not alignment across the junction of the two pieces of the flanking exons. It means that the two flanking exons are really exons but align separated may be because of an alternative splicing process. However, this cases should be tested to prove if really an alternative splicing have occurred.
- Alignment across junction: it means that almost all the query (about 100 pb) matches with the subject in only one region (it finds the concatenation of the two exon flanking sequences in a subject all together). In this case we can say that a U12 intron is located between two U12-intron-flanking exons. This category will be the most interesting to find novel U12 introns.
- One exon flank-left: in this case, only the left flanking exon sequence (about 50 pb) matches with the subject. The other may be an intron sequence that had been recognized as a U12 intron-flanking exon by GeneID and SGP. As it is an intron sequence, we can not find it in the EST and 'nr' databases.
- One exon flank-right: the same case as before but in the case of the right flanking exon.
- No hits found!: BLAST can not be able to find any good hit. It means that the two predicted regions as predicted U12-intron-flanking exons should be in reality intron sequences that are not represented in EST and 'nr' databases. In this group may be we can add those BLAST results that were skipped because of their low e-value and/or high number of gaps.
The parameters that we used to discriminate between significant and non-significant hits were an e-value under one, number of gaps under four and a percentage of identity higher than 97%.
In the next table there are the different categories of predictions after doing the BLASTN and BLASTX for the real and predicted U12 introns. You can follow the link if you want to see which predicted U12 introns correspond to each category:
- Alignment across junction: it means that almost all the query (about 100 pb) matches with the subject in only one region (it finds the concatenation of the two exon flanking sequences in a subject all together). In this case we can say that a U12 intron is located between two U12-intron-flanking exons. This category will be the most interesting to find novel U12 introns.
As it can be seen in the table, positive control files ('C' files) contain other categories than 'aligment across junction', that was what we expected. It may be due to that our supervisor and we have used different parameters to filter BLAST hits.
The next graphic is a comparison of the results of BLASTN and BLASTX, according with the classification of the predicted U12 introns ('P' file) in the different categories.

As we have explained before, predicted U12 introns contained in the 'alignment across junction' category are strong candidates to be novel U12 introns. To prove it, its nucleotide sequence should correspond with an intron in a database such as the UCSC Genome Browser. The UCSC Genome Browser provides a rapid and reliable display of any requested portion of genomes at any scale, together with dozens of aligned annotation tracks (known genes, predicted genes, ESTs, mRNAs, CpG islands, assembly gaps and coverage, chromosomal bands, mouse homologies, and more). Half of the annotation tracks are computed at UCSC from publicly available sequence data. The remaining tracks are provided by collaborators worldwide. Users can also add their own custom tracks to the browser for educational or research purposes.
We took some of the predicted U12 introns ('P' file) from the 'alignment across junction' category (from BLASTX and BLASTN) in order to determine if its position really corresponds with an intron position in the UCSC Genome Browser. Here we present some of the novel U12 introns that we found:
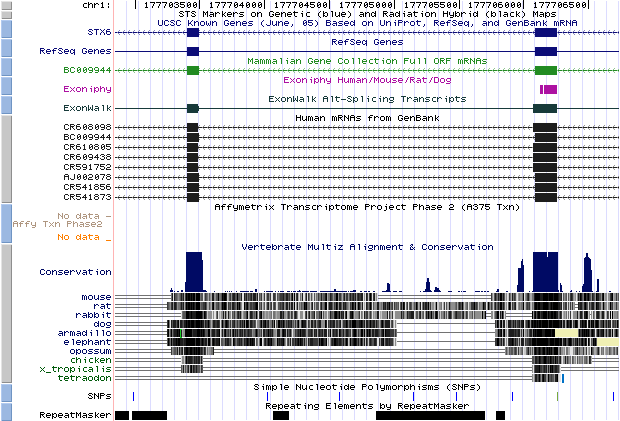
- chr1_1859|chr1|177703494|177706086|- (hit obtained with BLASTN against a dbEST):
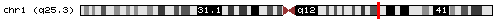
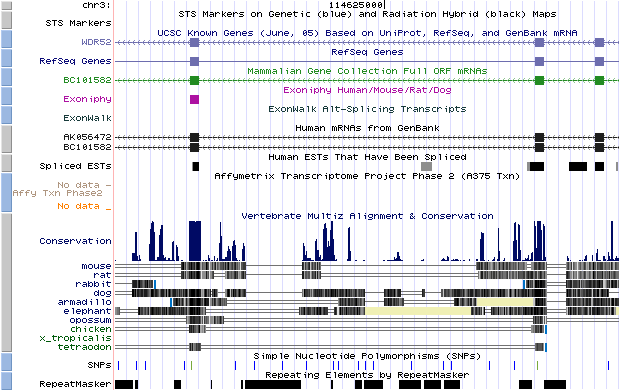
- chr3_2520|chr3|114621717|114627660|- (hit obtained with BLASTN against a dbEST):
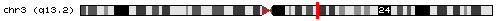
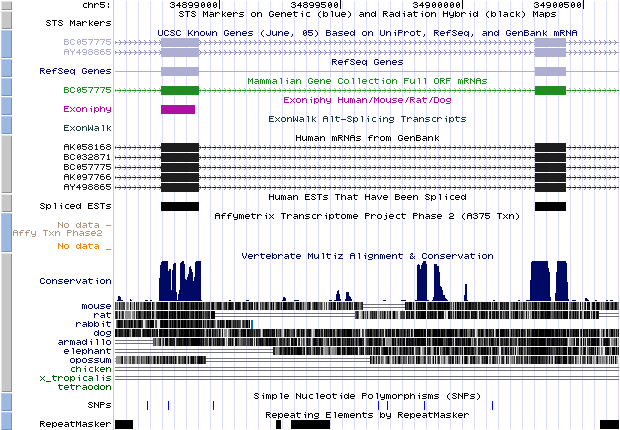
- chr5_259|chr5|34898917|34900298|+ (hit obtained with BLASTX against a 'nr' database):

- chrY_93|chrY|9239299|9239905|+ (hit obtained with BLASTX against a 'nr' database):


- chr3_2520|chr3|114621717|114627660|- (hit obtained with BLASTN against a dbEST):
If we look the start and the end position of the intron (thin grey line) we would appreciate that it corresponds with our predicted U12 intron, so it means that this predicted intron really exist in the database and we can affirm it is a U12 intron because its flanking exons had been predicted with Geneid_v1.2_u12 and SGP as U12 intron-flanking exons. We can also see where it is located in the chromosome.
At this point, more analysis with the UCSC Genome Browser should be done to locate and confirm all the predicted U12 introns classified in the 'Alignment across junction' category. Moreover, it would be interesting to compare if BLASTX and BLASTN obtain the same U12 introns or not. Another think that could be interesting is classifying U12 introns in ATAC or GTAG group.

|
HOME |

|