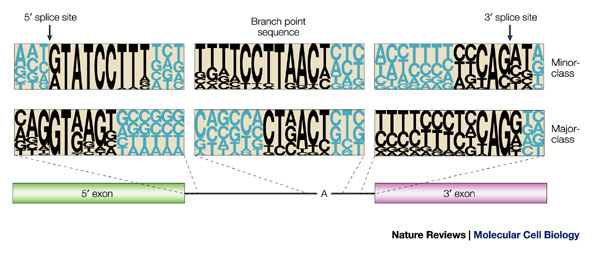
Figure 1. Consensus sequences of major-class and minor-class intons [2].

Figure 1. Consensus sequences of major-class and minor-class intons [2].
To process the pre-mRNA, the spliceosome complex is assembled in an ordered process involving five small nuclear ribonucleoprotein particles (snRNPs) and also several protein factors. The snRNAs used in the processing of U12 introns are U11, U12, U4atac, U6atac and U5. The latter is the same used in the processing of U2 introns; the other four are anologous to U1, U2, U4 and U6 used with the major class introns. Many proteins participate in the splicing reaction, some of them are snRNPs, whereas others are non-snRNPs factors. Many of the snRNP-associated proteins are shared by both spliceosomes [2]. Introns spliced by the U12 spliceosome coexist in the same gene with U2-type introns.
U12-type introns have been found in the majority of metazoan and in plants. They have not been found in fungi or in Caenorhabditis elegans, where they seem to have been lost. There is no evidence of this type of introns in protists [3].
Burge et al. [3] found conservation of U12-type introns between different classes of vertebrates. Conservation of the minor intron has also been reported between paralogous genes. Subtype switching between AT-AC and GT-AG U12 introns has been detected as well as conversion from U12 to U2 introns [3]. This conversion seems to be only one way, because of the more constrained sites in the U12-type intron [1]. Several hypotheses about U12 introns evolution have been proposed [2,3]. It seems that they share a commom ancestor, but it is not clear how they have evolved [2,3].
Since the novel type of introns was discovered, different works have been carried out to found U12 introns in different organisms. In 1998, Burge et al.[3] prepared a set of 60 genes in the human genome containing this type of introns. In 2001 Levine and Durbin [4] extended the set until 404 different genes, including some introns with termini that are different to the usual ones AT-AC or GT-AG.
The objective of this work is to enlarge a bit the knowledge of the evolution of the minor class introns, following the history of three genes with U12 introns across 13 species, looking for phenomena such as homologous introns, subtype switching, conversion of intron type or intron loss. A small analysis within paralogous genes has also been performed in order to extend our insight to this evolutionary history.