Simple Implementation of the BLAST algorithm
The objective of this project is to
develop a simple PERL implementation of the ungapped
BLAST algorithm for the comparison of
two protein (or DNA) sequences according to a specified
scoring matrix.
INTRODUCTION
A protein has one or more active sites that characterize
its biochemical function. Acceptance of mutations in the middle
of an active site is uncommon as they can disrupt
the protein function. On the other hand, the segments between
the active sites usually allow for more mutations, as they
play a role in giving the protein its shape but
not in its function.
Accordingly, when comparing two proteins to infer a potential
functional homology, we must take into account this variability.
It is for this reason that the standard way of performing these
searches is with local alignment algorithms. A local alignment of two
sequences is an alignment of any substring of one sequence with any
substring of the other sequence.
The local similarity of two sequences is defined as the highest
score obtained by any local alignment between the two sequences.
The Needleman-Wunsch dynamic programming algorithm for the global
alignment of two sequences can be modified to produce a local alignment.
This new algorithm is known as the Smith-Waterman algorithm (SW).
The SW algorithm is, however, very slow when searching a large amount of sequences.
A way of improving the running time of the comparison algorithm
is using 'heuristics'. This means that, although they can find the best
scoring alignment, there is no guarantee of doing so.
BLAST is one of those heuristics methods.
BLAST heuristic relies on the following two assumptions:
- The first is that most high-scoring alignments contain
one or more high-scoring pairs of W-letter substrings called words.
The value used for W is typically 3 for aminoacids (10 for nucleotides), but it can vary.
BLAST collects seed-alignments by identifying places in both
sequences where high-scoring word pairs occur.
- The second is that homologous sequences usually contain
gap-free alignments of considerable length. Accordingly, the
seed-alignments can be subsequently extended to form
gap-free alignment blocks.
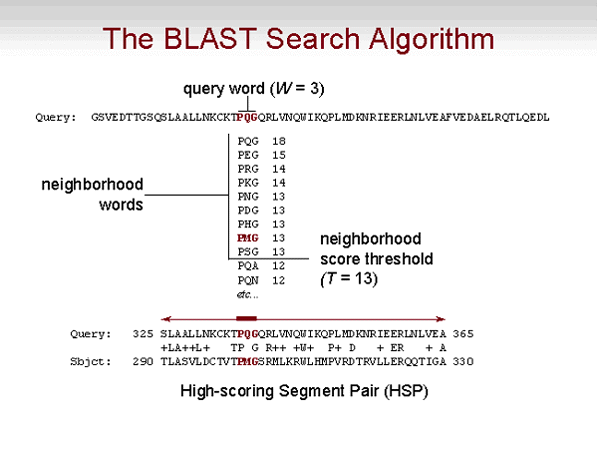
The BLAST algorithm can then be divided into the following steps:
- In the first step the query sequence is processed.
We consider a table of all the possible words
(for 3-letter words: 20*20*20=8000). To each
word we associate the list of positions in the
query sequence of all the words than give a high score when aligned with
the key word. This is called the query index.
The score is calculate according to the scoring matrix.
The exact 3-letter match is always high scoring, but note that other
matches are also possibly high.
- In the second step, we scan the target sequence. We consider all the 3-letter
words in the target sequence, and look them up in the query index, to obtain
the match position and score to the query. Initially one can consider exact matches.
In order to increase sensitivity, we can use a parameter T, and use all the words in the
target that score T or greater according to an specified substitution matrix.
- Each word hit is subsequently extended to the right and to the left to increase
the alignment's score, up to whatever position maximizes the overall
alignment score. Note that extensions beyond the first negative score are pointless.
Each alignment built in
this way is an high-scoring pair or HSP.
- The minimum score (S) for the
reported HSPs ( and the maximum allowed expected value (E) of the HSPs, but we will
not work with E values in this case )
can be specified by the user. HSPs that meet these criteria will be
reported by BLAST. Additionally, the user can also specify the maximum
number of reported 'found targets' (B) and HSPs (alignments) (V). Note
that there can be more than one alignment per target.
SUGGESTED WORK PLANNING
-
Write a perl program that reads
two protein sequences in fasta format and a scoring matrix, e.g.:
./simple_blast.pl seq1.fa seq2.fa blosum62
-
The program should hold in memory the sequences and the scoring matrix for
later manipulation.
-
Consider the first file to correspond to the query.
Modify the program above to add the processing of the query sequence, i.e.
build a query index for the query sequence:
For each 3-letter word in the query sequence, find all the
neighbouring words ( the word neighbourhood), that is, all the
3-letter words that only differ from the query word in
one letter and have a high score when compared to it. A score
is usually high when it is >= 11.
-
Modify the script to allow to introduce the length of the word
and the minimum score for the word nieghbourhood as parameters,
W and T, respectively, e.g.
./simple_blast.pl seq1.fa seq2.fa blosum62 W=4 T=10
-
Extend the program to add a function to scan the target sequence.
This function should search the target sequence word by word (note the word length
used = W) and use the table built from the query sequence to detect hits.
Store the found hits.
-
Extend the program to add a function that extend each one of the hits
found in the target sequence. The extension should stop before we
add an alignment that makes the score negative. Store all the alignments.
-
Extend the program to print the alignments sorted by score and
beyond a certain length that we can pass as a paramter, e.g.:
./simple_blast.pl seq1.fa seq2.fa blosum62 W=4 T=10 L=20
-
It is very important that the program is well documented
and for each function there is a clear description of what it does.
This documentation must be insde the program and can be in the
format of Perl comment using the symbol #.
The description of the project and the results obtained should be
clearly explained in a simple web site or in article format. It is
important that the work is presented in a way that is reproducible by
others. The construction of a web-server will be positively evaluated.

