RPS-BLAST 2.2.5 [Nov-16-2002]
Query= local sequence:
hg13_dna|geneid_v1.1_predicted_protein_2|1064_AA
(1063 letters)
Database: cdd.v1.60
10,013 PSSMs; 2,494,783 total columns
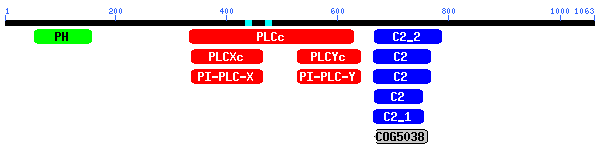
 .. This CD alignment includes 3D structure. To display structure, download
Cn3D! .. This CD alignment includes 3D structure. To display structure, download
Cn3D!
| |
PSSMs producing significant alignments: |
Score
(bits) |
E
value |
| |
 |
gnl|CDD|5336 |
cd00137, PLCc, Phospholipase C, catalytic domain; Phosphoinosi... |
284 |
4e-77 |
 |
gnl|CDD|133 |
smart00148, PLCXc, Phospholipase C, catalytic domain (part); d... |
174 |
4e-44 |
 |
gnl|CDD|134 |
smart00149, PLCYc, Phospholipase C, catalytic domain (part); d... |
167 |
5e-42 |
 |
gnl|CDD|951 |
pfam00388, PI-PLC-X, Phosphatidylinositol-specific phospholipa... |
144 |
5e-35 |
 |
gnl|CDD|950 |
pfam00387, PI-PLC-Y, Phosphatidylinositol-specific phospholipa... |
139 |
2e-33 |
 |
gnl|CDD|14861 |
cd00275, C2_2, Protein kinase C conserved region 2, subgroup 2... |
120 |
6e-28 |
 |
gnl|CDD|8952 |
smart00239, C2, Protein kinase C conserved region 2 (CalB); Ca... |
78.3 |
4e-15 |
 |
gnl|CDD|14771 |
cd00030, C2, Protein kinase C conserved region 2 (CalB); Ca2+-... |
75.1 |
3e-14 |
 |
gnl|CDD|9086 |
pfam00168, C2, C2 domain |
71.9 |
3e-13 |
 |
gnl|CDD|14862 |
cd00276, C2_1, Protein kinase C conserved region 2, subgroup 1... |
51.0 |
7e-07 |
| |
gnl|CDD|14168 |
COG5038, COG5038, COG5038, Ca2+-dependent lipid-binding protei... |
40.7 |
8e-04 |
 |
gnl|CDD|9087 |
pfam00169, PH, PH domain. PH stands for pleckstrin homology |
37.5 |
0.007 |
 |
gnl|CDD|5336,
cd00137, PLCc, Phospholipase C, catalytic domain; Phosphoinositide-specific
phospholipases C catalyze hydrolysis of phosphatidylinositol-4,5-bisphosphate
(PIP2) to D-myo-inositol-1,4,5-trisphosphate (1,4,5-IP3) and sn-1,2-diacylglycerol
(DAG). Both products function as second messengers in eukaryotic signal transduction
cascades. 1,4,5-IP3 triggers inflow of calcium from intracellular stores;
the membrane resident product DAG controls cellular protein phosphorylation
states by activating various protein kinase C isozymes. The enzyme comprises
2 regions (X and Y) connected via a linker which may contain inserted domains,
X and Y together form a TIM barrel-like structure containing the active site
residues. |
CD-Length = 298 residues, 100.0% aligned
Score = 284 bits (727), Expect = 4e-77
Query: 331 KVCQDMKQPLSHYFINSSHNTYLIEDQFRGPSDITGYIRALKMGCRSVELDVWDGPDNEP 390
Sbjct: 1 MVAQDMSKPLSHYFIPSSHNTYLTGKQVWGESSIEGYIQALKHGCRCVELDCWDGPDNEP 60
Query: 391 VIYTGHTMTSQIVFRSVIDIINKYAFFASEYPLILCLENHCSIKQQKVMVQHMKKLLGDK 450
Sbjct: 61 VVYHGHTFTTPIKLSEVLEAIKDFAFVTSPYPVILSLEDHCSPDQQAKMADSFKETFGDL 120
Query: 451 LYTTSPNVEESYLPSPDVLKGKILIKAKKLSSNCSGVEGDVTDEDEGAEMSQRMGKENME 510
Sbjct: 121 LYTPPTFSSLNVLPSPE-----QLKGKILLKGKKSGTYLDALEKEEGDSSQHSDSSESMS 175
Query: 511 QPNNVPVKRFQLCKELSELVSICKSVQFKEFQ-VSFQVQKYWEVCSFNE--VLASKYANE 567
Sbjct: 176 SEKKPSEKTHIRIAPESSELIGYQSLQWKDGETFTTESNQSLNIFSQSEYKVLLVKAIKE 235
Query: 568 NPGDFVNYNKRFLARVFPSPMRIDSSNMNPQDFWKCGCQIVAMNFQTPGLMMDLNIGWFR 627
Sbjct: 236 TPLKLVKTNQNYLLRVYPSGTRGDSSNYNPQIAWNAGVQIVALNFQTYGEGMQLNLGMFR 295
Query: 628 QNG 630
Sbjct: 296 ANG 298
 |
gnl|CDD|133,
smart00148, PLCXc, Phospholipase C, catalytic domain (part); domain X; Phosphoinositide-specific
phospholipases C. These enzymes contain 2 regions (X and Y) which together
form a TIM barrel-like structure containing the active site residues. Phospholipase
C enzymes (PI-PLC) act as signal transducers that generate two second messengers,
inositol-1,4,5-trisphosphate and diacylglycerol. The bacterial enzyme appears
to be a homologue of the mammalian PLCs. |
CD-Length = 145 residues, 92.4% aligned
Score = 174 bits (443), Expect = 4e-44
Query: 334 QDMKQPLSHYFINSSHNTYLIEDQFRGPSDITGYIRALKMGCRSVELDVWDGPDNEPVIY 393
Sbjct: 1 QDMSKPLSHYFINSSHNTYLTGKQLWGESSVEGYIQALKHGCRCVELDCWDGPDGEPVIY 60
Query: 394 TGHTMTSQIVFRSVIDIINKYAFFASEYPLILCLENHCSIKQQKVMVQHMKKLLGDKLYT 453
Sbjct: 61 HGHTFTLPIKLSEVLEAIKKFAFVTSPYPVILSLENHCSPDQQAKMAQMFKEIFGDLLYT 120
Query: 454 TSPNVEESYLPSPD 467
Sbjct: 121 PPTTSSLEYLPSPE 134
 |
gnl|CDD|134,
smart00149, PLCYc, Phospholipase C, catalytic domain (part); domain Y; Phosphoinositide-specific
phospholipases C. These enzymes contain 2 regions (X and Y) which together
form a TIM barrel-like structure containing the active site residues. Phospholipase
C enzymes (PI-PLC) act as signal transducers that generate two second messengers,
inositol-1,4,5-trisphosphate and diacylglycerol. The bacterial enzyme appears
to be a homologue of the mammalian PLCs. |
CD-Length = 117 residues, 100.0% aligned
Score = 167 bits (425), Expect = 5e-42
Query: 526 LSELVSICKSVQFKEFQVSFQVQKYWEVCSFNEVLASKYANENPGDFVNYNKRFLARVFP 585
Sbjct: 1 LSELVSYCAPVKFRSFELAEEKNPFYEMSSFSETKAKKLLEKAPTDFVRYNQRQLSRVYP 60
Query: 586 SPMRIDSSNMNPQDFWKCGCQIVAMNFQTPGLMMDLNIGWFRQNGNCGYVLRPAIMR 642
Sbjct: 61 KGTRVDSSNYNPQVFWNHGCQMVALNFQTPDKAMQLNQGMFRANGGCGYVLKPDFLR 117
 |
gnl|CDD|951,
pfam00388, PI-PLC-X, Phosphatidylinositol-specific phospholipase C, X domain.
This associates with pfam00387 to form a single structural unit. |
CD-Length = 145 residues, 91.7% aligned
Score = 144 bits (365), Expect = 5e-35
Query: 335 DMKQPLSHYFINSSHNTYLIEDQFRGPSDITGYIRALKMGCRSVELDVWDGPDNEPVIYT 394
Sbjct: 1 DMSIPLSHYFISSSHNTYLTGKQLWGKSQVESYRQQLDHGCRCVELDCWDGPDDEPIIYH 60
Query: 395 GHTMTSQIVFRSVIDIINKYAFFASEYPLILCLENHCSIKQQKVMVQHMKKLLGDKLYTT 454
Sbjct: 61 GGTFTLEIKLKDVLEAIKDFLFKTSPYPIILSLENHCNSDQQRKMAKYFEEIFGDYLLTK 120
Query: 455 SPNVEESY-LPSPD 467
Sbjct: 121 -PLDSLTTKLPSLK 133
 |
gnl|CDD|950,
pfam00387, PI-PLC-Y, Phosphatidylinositol-specific phospholipase C, Y domain.
This associates with pfam00388 to form a single structural unit. |
CD-Length = 118 residues, 100.0% aligned
Score = 139 bits (351), Expect = 2e-33
Query: 525 ELSELVSICKSVQFKEFQVSFQVQKYWEVCSFNEVLASKYANENPGDFVNYNKRFLARVF 584
Sbjct: 1 ELSNLVNYIQSIKFRSFSLPTEKNTSYEMSSFSERKAKQLLKESPIEFVKHNKRQLSRVY 60
Query: 585 PSPMRIDSSNMNPQDFWKCGCQIVAMNFQTPGLMMDLNIGWFRQNGNCGYVLRPAIMR 642
Sbjct: 61 PKGTRFDSSNFMPQPFWNAGCQMVALNFQTSDLPMQINLGMFEYNGGSGYLLKPPFLR 118
 |
gnl|CDD|14861,
cd00275, C2_2, Protein kinase C conserved region 2, subgroup 2; C2 Ca2+-binding
motif present in phospholipases, protein kinases C, and synaptotagmins (amongst
others); some PKCs lack calcium dependence. Particular C2s appear to bind
phospholipids, inositol polyphosphates,and intracellular proteins. Two distinct
C2 topologies generated by permutation of the sequence with respect to the
N- and C-terminal beta strands are seen. In this subgroup, containing phospholipases
C and D( PLC-1, PLD) and specific protein kinases C (PKC) subtypes, the C-terminal
beta strand occupies the position of what is the N-terminal strand in subgroup
1. |
CD-Length = 129 residues, 99.2% aligned
Score = 120 bits (303), Expect = 6e-28
Query: 664 LHIKIISGQNFPKPKGS--GAKGDVVDPYVYVEIHGIPADCAEQRTKTVHQNGDAPIFDE 721
Sbjct: 2 LKVRIIEAQQLPKTDKSLDGKSKSTLDPYVTVEIDGVDA--RIGRTKVIQNNGFNPVWNE 59
Query: 722 SFEFQINLPELAMVRFVVLDDDYI-GDEFIGQYTIPF-ECLQTGYRHVPLQSLTGEVLAH 779
Sbjct: 60 EFEFPLAHPDLAFIRFTVKDSDPIGGDDFIGNATIPVDEVGKQGYRWIPLLDMNGEQLPF 119
Query: 780 ASLFVHVAIT 789
Sbjct: 120 SKLFVKIQLE 129
 |
gnl|CDD|8952,
smart00239, C2, Protein kinase C conserved region 2 (CalB); Ca2+-binding
motif present in phospholipases, protein kinases C, and synaptotamins (among
others). Some do not appear to contain Ca2+-binding sites. Particular C2s
appear to bind phospholipids, inositol polyphosphates, and intracellular
proteins. Unusual occurrence in perforin. Synaptotagmin and PLC C2s are permuted
in sequence with respect to N- and C-terminal beta strands. SMART detects
C2 domains using one or both of two profiles. |
CD-Length = 101 residues, 100.0% aligned
Score = 78.3 bits (192), Expect = 4e-15
Query: 663 LLHIKIISGQNFPKPKGSGAKGDVVDPYVYVEIHGIPADCAEQRTKTVHQNGDAPIFDES 722
Sbjct: 1 TLTVKIISARNLPPKDKGGK----SDPYVKVSLDGDPREKK--KTKVVKNTLN-PVWNET 53
Query: 723 FEFQINLPELAMVRFVVLDDDYIG-DEFIGQYTIPFECLQTGYRHVPL 769
Sbjct: 54 FEFEVPPPELSELEIEVYDKDRFSRDDFIGQVTIPLSDLLLGGRHEKL 101
 |
gnl|CDD|14771,
cd00030, C2, Protein kinase C conserved region 2 (CalB); Ca2+-binding motif
present in phospholipases, protein kinases C, and synaptotagmins (among others).
Some do not appear to contain Ca2+-binding sites. Particular C2s appear to
bind phospholipids, inositol polyphosphates,and intracellular proteins. Synaptotagmin
and PLC C2s are permuted in sequence with respect to N- and C-terminal beta
strands. |
CD-Length = 105 residues, 100.0% aligned
Score = 75.1 bits (184), Expect = 3e-14
Query: 663 LLHIKIISGQNFPKPKGSGAKGDVVDPYVYVEIHGIPADCAEQRTKTVHQNGDAPIFDES 722
Sbjct: 1 RLTVKIIEARNLPPKDKKGT----SDPYVKVSLGGDKK--QKKKTKVV-KKTLNPVWNET 53
Query: 723 FEFQINLPELAMVRFVVLDDDYIG-DEFIGQYTIPFECL----QTGYRHVPL 769
Sbjct: 54 FTFEVPPPEESSLVIEVYDYDKFSRDDFIGEVTIPLSELLLDGKEGDRWFPL 105
CD-Length = 85 residues, 100.0% aligned
Score = 71.9 bits (176), Expect = 3e-13
Query: 664 LHIKIISGQNFPKPKGSGAKGDVVDPYVYVEIHGIPADCAEQRTKTVHQNGDAPIFDESF 723
Sbjct: 1 LRVTVIEAKNLPPKDKNGK----SDPYVKVSLGGQKKDT--KKTKVIKNTLN-PVWNETF 53
Query: 724 EFQINLPELAMVRFVVLDDDYIG-DEFIGQYT 754
Sbjct: 54 TFEVPPPELAELRIEVYDYDRFGKDDFIGEVS 85
 |
gnl|CDD|14862,
cd00276, C2_1, Protein kinase C conserved region 2, subgroup 1; C2 Ca2+-binding
motif present in phospholipases, protein kinases C, and synaptotagmins (amongst
others); some PKCs lack calcium dependence. Particular C2s appear to bind
phospholipids, inositol polyphosphates,and intracellular proteins. Two distinct
C2 topologies generated by permutation of the sequence with respect to the
N- and C-terminal beta strands are seen. In this subgroup, containing synaptotagmins,
specific protein kinases C (PKC) subtypes and other proteins, the N-terminal
beta strand occupies the position of what is the C-terminal strand in subgroup
2. |
CD-Length = 124 residues, 75.0% aligned
Score = 51.0 bits (122), Expect = 7e-07
Query: 662 QLLHIKIISGQNFPKPKGSGAKGDVVDPYVYVEIHGIPADCAEQRTKTVHQNGDAPIFDE 721
Sbjct: 15 GQLTVVIIKARNLPPMDKNGL----SDPYVKVYLLPDGKKKKKKKTK-VKRKTLNPVFNE 69
Query: 722 SFEFQINLPELAMVR--FVVLDDDYIG-DEFIGQYTIP 756
Sbjct: 70 TFVFDVPPEELADRSLQITVWDYDRFSRNDFIGEVVIG 107
gnl|CDD|14168, COG5038, COG5038, Ca2+-dependent lipid-binding protein, contains C2 domain [General function prediction only]
CD-Length = 1227 residues, only 7.5% aligned
Score = 40.7 bits (95), Expect = 8e-04
Query: 664 LHIKIISGQNFPKPKGSGAKGDVVDPYVYVEIHGIPADCAEQRTKTVHQNGDAPIFDESF 723
Sbjct: 1042 LTIMLRSGENLP----SSDENGYSDPFVKLFLNE-----KSVYKTKVVKKTLNPVWNEEF 1092
Query: 724 EFQINLPELAMVRFVVLDDDYIG-DEFIGQYTIPFECLQTG 763
Sbjct: 1093 TIEVLNRVKDVLTINVNDWDSGEKNDLLGTAEIDLSKLEPG 1133
 |
gnl|CDD|9087, pfam00169, PH, PH domain. PH stands for pleckstrin homology.
|
CD-Length = 100 residues, 97.0% aligned
Score = 37.5 bits (86), Expect = 0.007
Query: 53 EGSELKKVRSNSRIYH-RYFLLDADMQSLRWEPSKKDSEKAKIDIKSIKEVRTGKNTDIF 111
Sbjct: 4 EGWLLKKSTVKKKRWKKRYFFLFNDVLIYYKDKKKSYEPKGSIPLSGC-SVEDVPDSEF- 61
Query: 112 RSNGISDQISEDCAFSVIYGENYESLDLVANSADVANIWVTGLRYLIS 159
Sbjct: 62 ---------KRPNCFQLRSRDGKETFILQAESEEERQDWIKAIQSAIR 100
|

 .. This CD alignment includes 3D structure. To display structure, download
Cn3D!
.. This CD alignment includes 3D structure. To display structure, download
Cn3D!