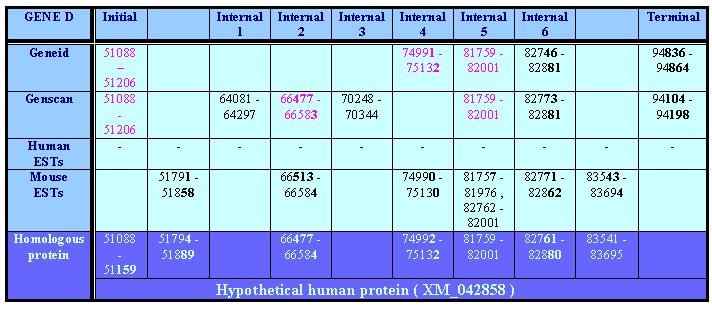

It is in forward strand and predicted by the two programs. Geneid precists one initial, three internals and a terminal exon, while genscan predicts one initial, five internals amd a terminal exon. Genscan also predicts a promotor and a polyA, which we won't use.
There is just one human EST in the region limitated by the predictions, but it doesn't reinforce any exon. Despite this, it has a length of 800 nucleotides, from which a 704 nucleotide region match with a socre of 99%. As it doesn't reinforce any prediction and it has 100 unaligned nuclotides, we don't consider it much informative. What we do find reinforcing our data is mouse ESTs.
With the gene from the genscan we obtain as a result an unknown protein (protein XM_042858 for MGC:15763) with a similarity of 73% in 204 of the 328 aminoacides predicted. With the gene from the geneid we obtain a 72% of similarity with the same unknown protein, matching 213 of 222 predicted aminoacids. We will analyze every exon, but in this case, as we don't have any human EST reinforcig, we will mostly trust the genepredictions to delimitate exactly the exons.
But the last exon predicted by the tblastn with the homologous protein is supported by a mouse EST. We consider this exon as good, because we search it manually in the geneid predictions. We search for a terminal exon coinciding with the one predicted with the homologous protein. We find one with good score and matching, so we select it.
This can be visulized in GhostView format.
Concerning the promotor sites, we do the same as in the previous case and obtain this view of the possible promotors.