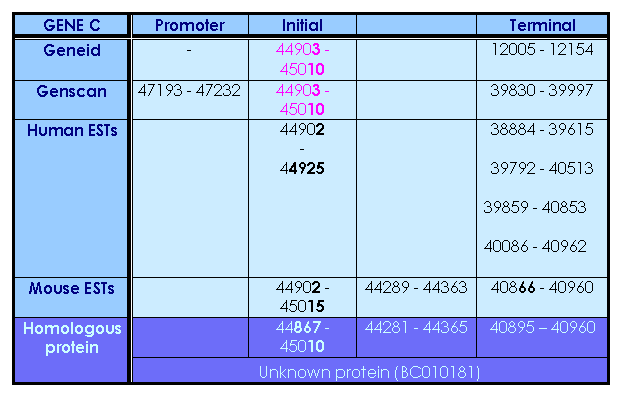

This gene is in reverse strand. Both programs predict 2 exons: an initial one, identically predicted in the two programs, and a terminal, which differes a lot between them. Furthermore, genscan predicts a promotor region, but, as has been comented previously, we won't take count of this. We will try to reinforce this initial prediction with the later information we obtain.
We run a blastp against the two predicted proteins (from genscan and geneid) and obtain the same results: only 36 aminoacids matched with the same score (99%) and with the same protein. As these two proteins had only in common the initial exon, we decide to do the blastp only with this exon, and find a match with an unknown protein (protein for MGC:20197), obtained from a cDNA library preparation. We obtain a 100% of similarity in 36 of the 38 predicted aminoacids in the initial exon.
Putting this information altogether:
This can be visulized in GhostView format.
So, we can conclude that the initial exon is the predicted by both programs and accept the two exons predicted by the tbalstn with the homologous protein. As terminal exon we finally accept the last exon predicted by the homologous protein.
Once we have this, as the initial exon is confirmed, we select the upstream region with the ss program (nucleotides 45010 to 47250) and run it against the TRANSFAC server to predict the promoter region. We obtain an output file which we convert to gff format (so that we can convert it into PostScript format and visualize the results with the GhostView) with this comand:
grep "^ V\$.*" gen2geneid.promotor.txt | gawk '{print "AC090948", $1, "prom", $3, $3+length($10), $6, ".",".","gen2geneid"}' > gen2genid.prom.gff
And we visualize the results.